This page was last revised for version 5.0.0.
Fully automated tracing routines can be accessed through the Auto-trace menu in the main dialog.
Grayscale Images
Grayscale images are processed using the Auto-trace › Grayscale Image… command for images already loaded or Auto-trace › Grayscale Image File… to load from disk (Big data support).
This command reconstructs neuronal structures directly from grayscale (intensity) images without requiring binarization. It uses the Gray-Weighted Distance Transform (GWDT) algorithm combined with Fast Marching and hierarchical pruning, following an APP2-like strategy. The method is particularly effective for fluorescence microscopy images with bright foreground structures on dark backgrounds.
You can follow these instructions using File › Load Demo Dataset… and choosing the Drosophila OP neuron, a Drosophila olfactory projection neuron and respective ground truth reconstruction.
Input Image
Grayscale Image (Mandatory) The intensity image to be traced (2D or 3D). Either the image currently loaded in SNT, or a Secondary image layer. Should contain bright neuronal structures on a dark background.
Soma/Root Detection
The algorithm requires a starting point (seed) to begin tracing. Several strategies are available:
-
None. Use auto-detection Automatically detects the soma/root at the thickest and brightest region using a combined Euclidean Distance Transform (EDT) and intensity scoring approach. This strategy finds the most prominent structure in the image, which typically corresponds to the cell body (see Soma Detection). No ROI is required
-
Area ROI around soma: One tree per primary neurite An area ROI delineates the soma boundary. Creates separate trees for each neurite exiting the soma, rooting them at the soma perimeter. Best for complex morphologies where multiple primary branches emerge from the cell body and individual neurites need to be analyzed separately
-
Single tree rooted at ROI centroid The ROI marks the soma location. Creates a single tree rooted at the geometric centroid of the ROI. Suitable for simple morphologies or when the entire arbor should be represented as one connected structure
-
Single tree rooted at ROI weighted centroid Similar to the above, but roots the tree at the intensity-weighted centroid of nodes traced within the soma region, rather than the ROI’s geometric center. More accurate when the ROI encompasses the entire soma but may not be precisely centered
When working with 3D images, the Active plane only option restricts the ROI to its specific Z-slice, preventing the algorithm from considering roots at other depths.
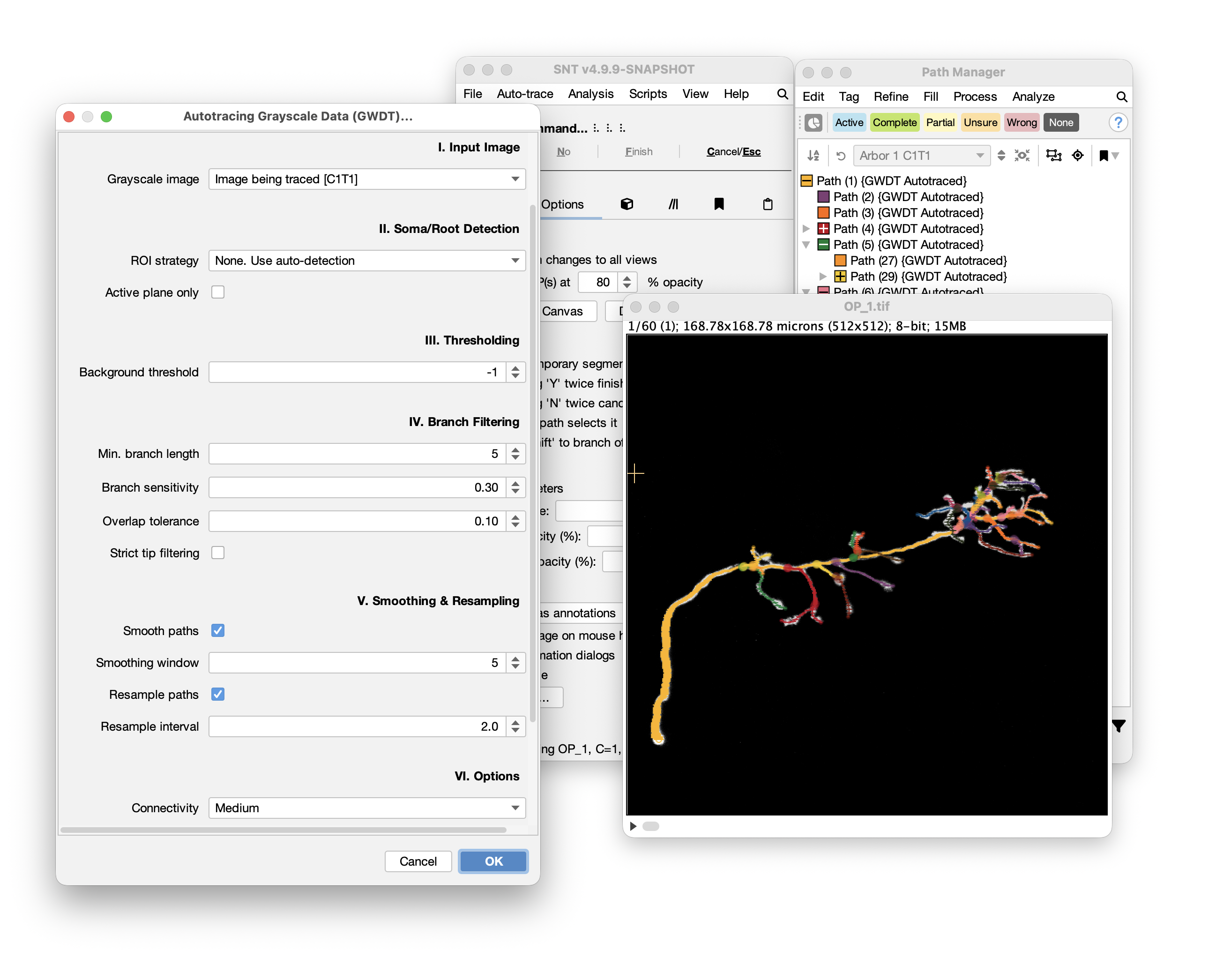
Thresholding
Background threshold Defines the intensity cutoff below which pixels are considered background and excluded from tracing. Lower values trace more structures (including noise), while higher values trace less (potentially missing dim neurites).
- Use
-1(default) for automatic threshold estimation using image statistics - Adjust manually if auto-detection includes too much noise or misses dim structures
- Valid range: -1 (auto) to the image maximum intensity
Branch Filtering
These parameters control which branches are retained in the final reconstruction:
Min. segment length The minimum intensity-weighted length threshold for branches to be included. This metric accumulates normalized pixel intensities along each branch path (rather than measuring spatial distance), so brighter branches accumulate length faster than dim ones. Branches below this threshold are pruned as noise. Increase to remove small spurious branches; decrease to retain finer details. Default: 5.0
Connectivity Defines the neighborhood connectivity used during tracing:
- Low (6-connected): Only axis-aligned neighbors
- Medium (18-connected): Axis-aligned plus face-diagonal neighbors (default)
- High (26-connected): All 26 voxel neighbors including edge and corner diagonals
Higher connectivity produces smoother paths through the image but may trace through gaps more liberally.
Post-processing
Terminal leaf pruning Removes short terminal branches that represent noise or incomplete tracing. Terminal branches are pruned if they substantially overlap with their parent branch (>90% coverage).
Smoothing Applies a moving average filter to reduce zigzag artifacts in traced paths. The window size determines how many adjacent nodes are averaged together. Larger windows create smoother paths but may lose fine details.
Resampling Redistributes nodes along paths at regular intervals to standardize node density. The step size (in voxels) determines spacing between resampled nodes. Useful for ensuring consistent spatial sampling across the reconstruction.
About the Algorithm
SNT’s grayscale autotracing has been implemented with substantial re-engineering/improvements:
-
Full-resolution tracing Works directly on original image data without mandatory downsampling, preserving fine details
-
Native bit-depth support Handles 8-bit, 16-bit, and 32-bit images without conversion, maintaining full dynamic range
-
Big data support with scalable architecture Automatically adapts to image size using optimized storage strategies: fast in-memory processing for typical images, memory-efficient sparse storage for larger images with sparse signal, or disk-backed processing for TB datasets that exceed available RAM
-
Automatic soma detection Locates the soma automatically using combined EDT × intensity scoring, eliminating the need for manual seed point placement (see Soma Detection)
-
Optimized parameters Default settings have been tuned and validated across SNT’s demo datasets, and are expected to work well out-of-the-box
For technical implementation details and references, see the SNT technical notes.
Soma Detection
The Auto-trace › Detect Soma… command provides automated detection of cell bodies in neuronal images. This is a standalone tool that can be used before autotracing to identify soma locations, or independently to annotate soma positions and sizes.
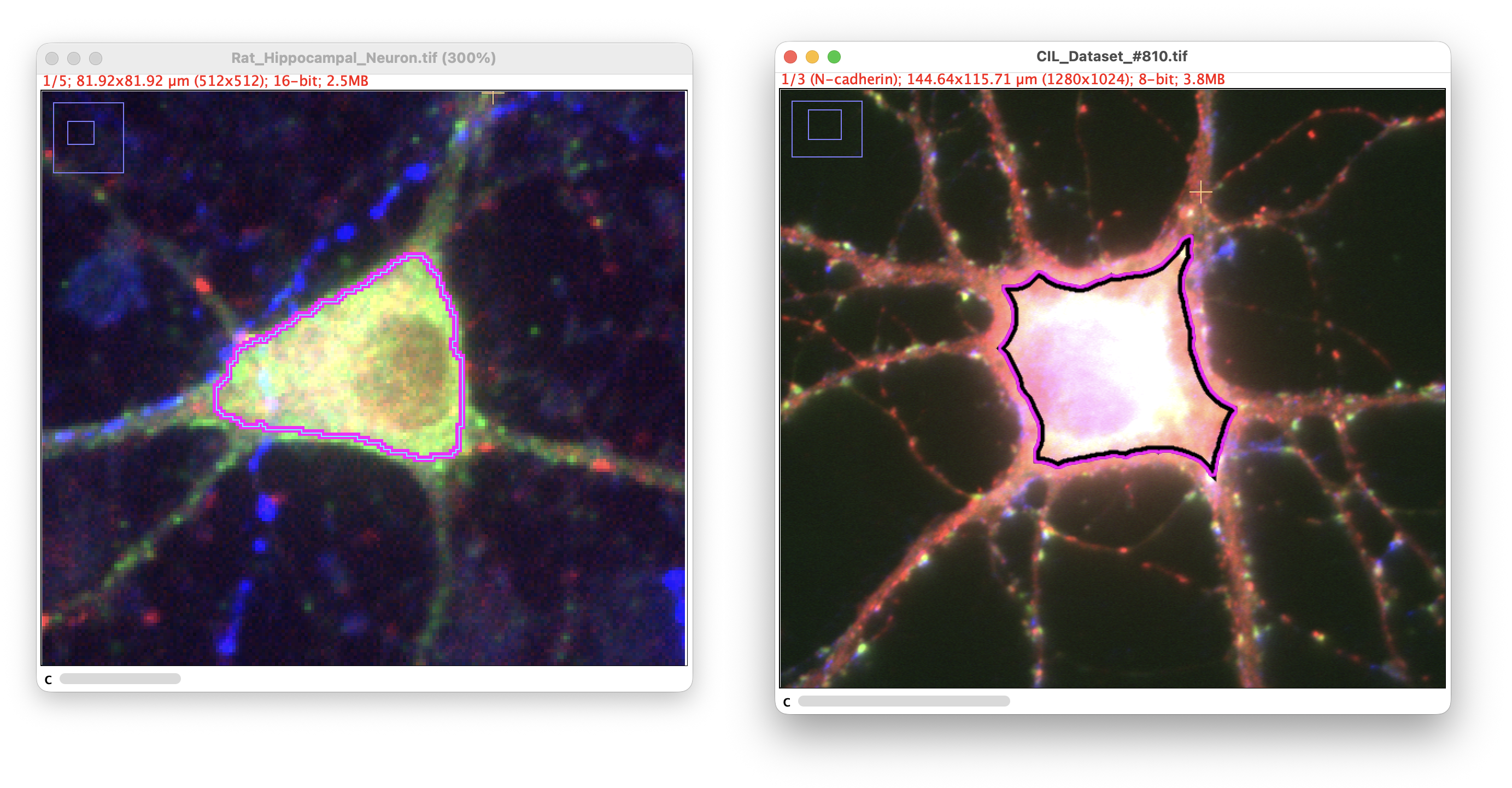
Automated soma detection: ROI countour (Cultured hippocamapl neurons: Neuronal receptors/Synaptic labeling demo datasets)
Detection Strategy
The algorithm uses a combined EDT-intensity approach to identify somas:
- Euclidean Distance Transform (EDT) identifies the thickest structures in the image. Since somas are typically the thickest part of neurons, they have the highest EDT values
- Intensity scoring weights the EDT by pixel intensity to favor bright structures
- The product of EDT × intensity identifies regions that are both thick and bright, typical characteristics of well-labeled cell bodies
This dual approach is more robust than using intensity alone because it effectively filters out hot pixels and imaging artifacts, which are bright but thin (low EDT values).
Detection Scope
Brightest/largest soma only Detects the single most prominent soma in the image—the structure with the highest EDT × intensity score. Use this for images containing a single neuron or when you want to identify the primary cell body.
All somata in image Detects multiple somas by finding local maxima in the EDT map. Each local maximum represents a thick structure that likely corresponds to a cell body. The Min. radius parameter filters out small detections (debris, hot pixels) by discarding structures below the specified radius.
Threshold
The intensity threshold determines which pixels are considered foreground during detection. Use -1 (default) to automatically compute the threshold using Otsu’s method. Manual adjustment may be needed for images with uneven illumination or weak signal.
Output Types
Results can be exported in several formats:
-
Single-node path Creates a path in SNT’s Path Manager consisting of a single node at the soma center. The node radius is estimated from the distance transform. Useful for marking soma locations in reconstructions
-
Point ROI Places a point marker at the detected soma center. Simplest output for marking locations
-
Area ROI Creates a contour ROI outlining the soma boundary, determined by flood-filling from the center at the detection threshold. Useful for measuring soma size or creating exclusion regions
-
Circular ROI Draws a circle centered on the soma with radius estimated from the distance transform. Approximates soma size when the cell body is roughly spherical
Segmented Images
Segmented (binary or thresholded) images are processed using skeleton-based tracing, which converts the image topology into neuronal tree structures. This approach is suitable for images with clear foreground/background separation where structures have already been segmented through thresholding or other preprocessing methods.
The algorithm works by first skeletonizing the segmented image to extract its topological structure, then converting the resulting skeleton graph into neuronal trees. Segmented images are processed using the Auto-trace › Segmented Image… command when the image to be processed is already open or Auto-trace › Segmented Image File…, if the image is yet to be loaded from disk.
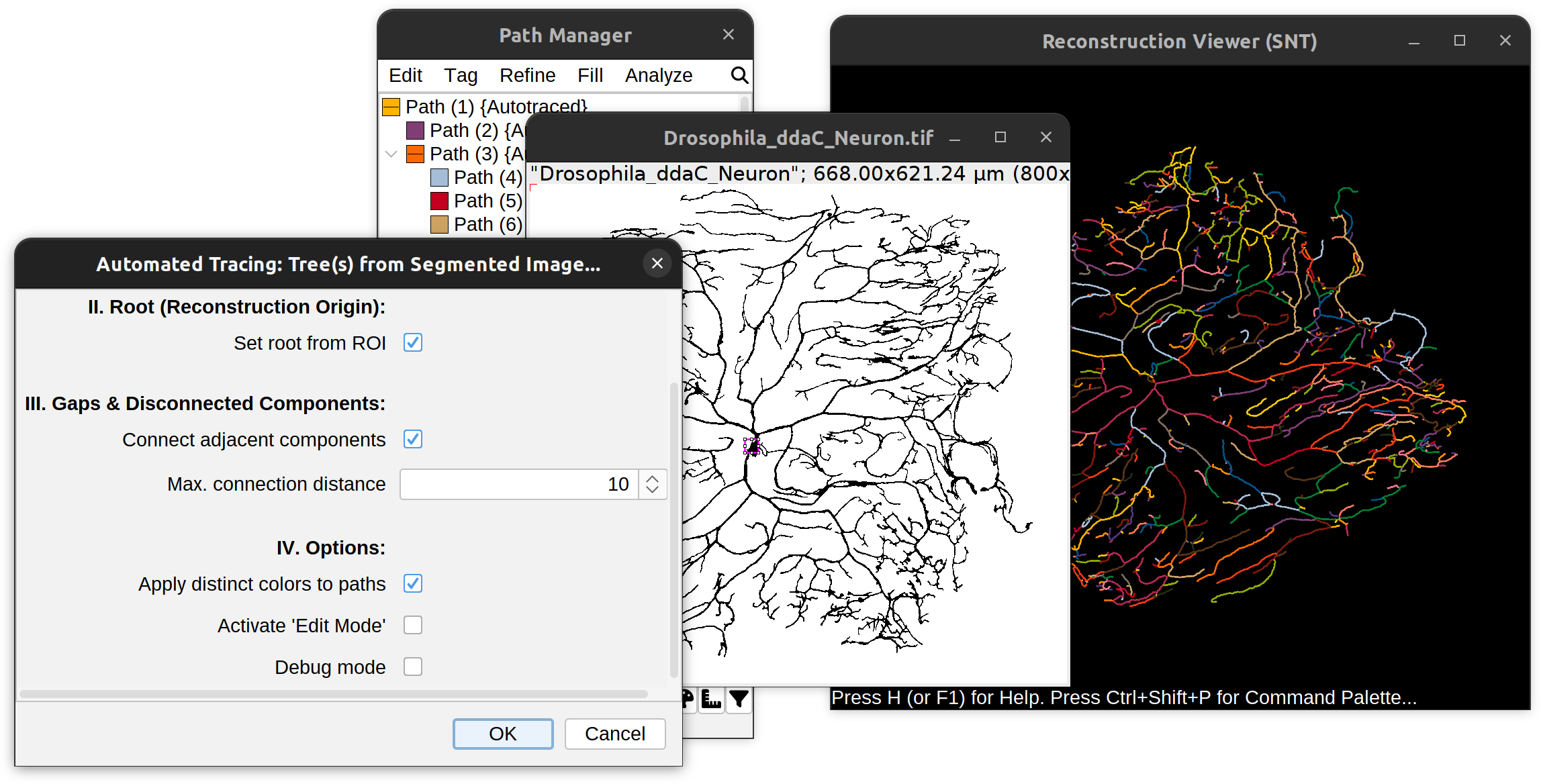
You can follow these instructions using File › Load Demo Dataset… and choosing the Drosophila ddaC neuron (Autotrace demo). It will load a binary (thresholded) image of a Drosophila space-filling neuron (ddaC) displaying autotracing options for automated reconstruction.
Input Image(s)
Full automated tracing of segmented (thresholded) images requires two types of inputs:
-
Segmented Image (Mandatory) The pre-processed image from which paths will be extracted. It will be skeletonized by the extraction algorithm. If thresholded (recommended), only highlighted pixels are considered, otherwise all non-zero intensities in the image will be considered for extraction.
-
Intensity Image (Optional) The original (un-processed) image used to resolve any possible loops in the segmented image using brightness criteria. Required for intensity-based loop resolution strategies (see below).
Soma/Root Detection
Root detection typically uses an area ROI to mark the root of the arbor on the image. With neurons, this typically corresponds to an area ROI marking the soma. Several strategies are possible:
-
Place root on ROI’s simple centroid Attempts to root all primary paths intersecting the ROI at its geometric centroid (the centroid of the ROI’s contour). Typically used when 1) multiple paths branch out from the soma, 2) the ROI accurately defines the contour of the soma, and 3) somatic segments are expected to be part of the reconstruction
-
Place root on ROI’s weighted centroid Similar to the above, but places the root at the centroid of all foreground voxels contained by the ROI, rather than the ROI’s geometric center. Useful when the ROI contour is imprecise but still encloses the relevant structure
-
Place roots along ROI’s edge Attempts to root all primary paths intersecting along the perimeter of the ROI. As above, this option is typically used when multiple paths branch out from the soma and the ROI defines the contour of the soma, but somatic segments are not expected to be included in the reconstruction. Most accurate strategy for complex topologies
-
ROI marks a single root Assumes a polarized morphology (e.g., apical dendrites of a pyramidal neuron), in which the arbor being reconstructed has a single primary branch at the root. Under this option, the ‘closest’ end-point (or junction point) contained by the ROI becomes the root node. In this case the ROI does not need to reflect an accurate contour of the soma
-
None. Ignore any ROIs If no ROI exists an arbitrary root node is used
When parsing 3D images, toggling the Active plane only checkbox clarifies that the root(s) marked by the ROI occurs at the active ROI’s Z-plane. This ensures that other possible roots above or below the ROI will not be considered.
Loop Resolution
Skeletonized images often contain spurious loops (cycles) that must be broken to produce valid tree structures. Several strategies are available:
-
Dimmest branch Removes the branch with the lowest average intensity in the loop. Requires an intensity image. Best when signal intensity correlates with structural importance
-
Dimmest voxel Cuts the loop at the single dimmest pixel. Requires an intensity image. More precise than dimmest branch but may be sensitive to noise
-
Shortest branch Removes the shortest branch (the skeletonized path between junctions) in the loop. Fast and suitable for simple artifacts, but may inadvertently break main structures
-
Shortest segment Removes the shortest edge(s) using a maximum spanning tree algorithm. Preserves the longest continuous paths in the structure. Suitable for simple loops where geometric length indicates “importance”
-
Furthest from root Removes edges most distant from the root/soma. Preserves proximal structure closer to the cell body. Requires a root-defining ROI. Only available when an ROI has been provided. Useful when loops near the soma are expected to be more meaningful/important than distal ones
-
Peripheral segments Removes edges with the lowest betweenness centrality (i.e., edges that lie on fewer shortest paths through the graph). Preserves the main backbone/trunk of the structure regardless of geometric length. Recommended for complex structures where topological importance matters more than geometry
NB: If an intensity-based strategy is selected but no intensity image is provided, the algorithm automatically falls back to Peripheral segments.
Gaps and Disconnected Components
It is almost impossible to segment structures into a single, coherent volume. These options define how gaps in the segmentation should be handled.
-
Settings for handling small components These options determine whether the algorithm should ignore segments (connected components that are disconnected from the main structure) that are too small to be of interest. Disconnected segments with a cable length below the specified length threshold will be discarded by the algorithm.
Increase this value if too many isolated branches are being created. Decrease it to bias the extraction for larger, contiguous structures. -
Settings for handling adjacent components If the segmented image is fragmented into multiple components (e.g., due to ‘beaded’ labeling): Should the algorithm attempt to connect nearby components? If so, what should be the maximum allowable distance between disconnected components to be merged?
You should increase Max. connection distance if the algorithm produces too many gaps. Decrease it to minimize spurious connections.
NB: When components are bridged, new loops may be introduced. These are automatically resolved using the selected loop resolution strategy.
Use Path Orientation (hold O) to verify path orientations. Hold F to temporarily hide all annotations.
Additional Options
Additional processing options are available in the dialog:
- Replace existing paths Whether any existing paths in the Path Manager should be discarded before adding newly traced structures
- Apply distinct colors Whether each traced tree should be assigned a unique color to facilitate visual distinction
- Activate ‘Edit Mode’ Whether SNT’s Edit Mode should be automatically activated after tracing completes, allowing immediate refinement of results
- Prune single-node paths Whether single-point paths without children should be filtered out from the final reconstruction
For technical implementation details including the skeletonization algorithm and graph conversion methods, see the SNT technical notes.
