The content of this page has not been vetted since shifting away from MediaWiki. If you’d like to help, check out the how to help guide!
Automatic Particle counting
Automatic particle counting can be done if the image does not have too many individual particles touching. Manual particle counting can be done using the Multi-point Tool.
Segmentation, or the ability to distinguish an object from its background, can be a difficult issue to deal with. Once this has been done, however, the object can then be analyzed.

RAW Threshold Watershed “AnalyzeParticles”
Setting a threshold
5.1.1.1 Manual thresholding
Automatic particle analysis requires a “binary”, black and white, image. A threshold range is set to tell the objects of interest apart from the background. All pixels in the image whose values lie under the threshold are converted to black and all pixels with values above the threshold are converted to white, or vice-versa.
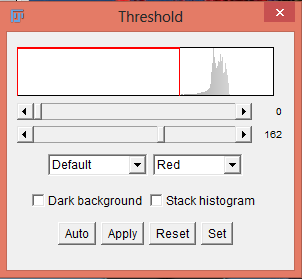
There are several ways to set thresholds. Monochrome images are most simply thresholded via the menu command Image › Adjust › Threshold. The threshold can be set using the slider bars. The pixels within the threshold range are displayed in red. When you are satisfied with the threshold settings, you can then hit Apply. This will permanently apply the threshold settings and convert the image to binary. You have different options for setting a manual threshold. The drop-down menu set to Default allows you to choose between Default and 15 other threshold techniques. The drop-down menu set to Red allows you to choose between a red on white color scheme, a black on white color scheme, or an over and under color scheme. The Dark Background box will flip the foreground color with the background color. You can also choose to check the Stack histogram box to produce a histogram for an entire stack.
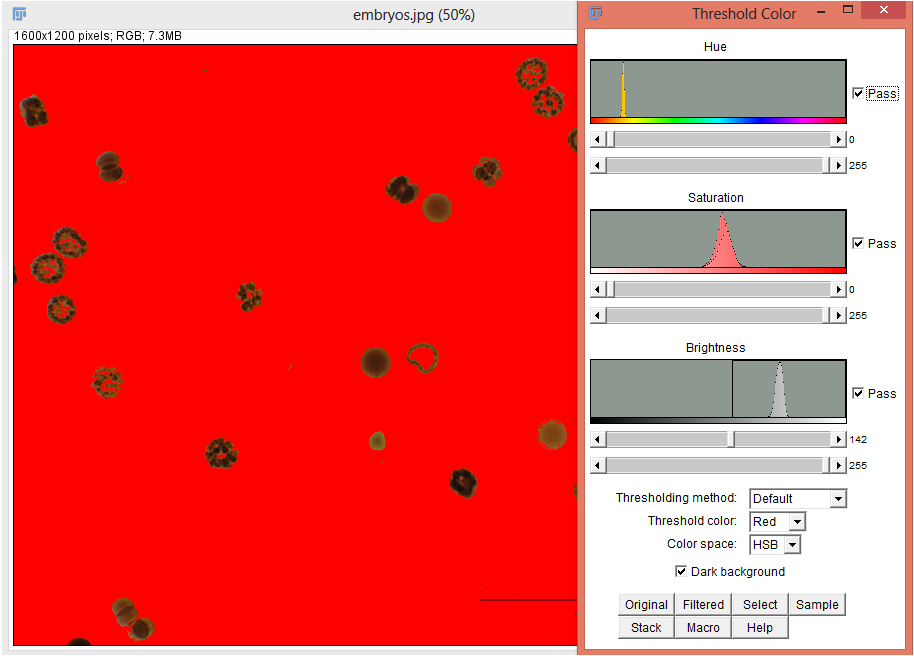
For color images, setting the threshold is done with the command sequence Image › Adjust › Color Threshold…. The Thresholding method option allows you to choose a thresholding techniqe other than the default. The Threshold color option allows you to choose between Red, White, Black, or B&W as the thresholding color. The Color space option allows you to choose between HSB, RGB, Lab, and YUV. The background of the thresholded image can be made light or dark. The image can be converted to a binary image via the menu command Image › Type › 8-bit.
Automatic thresholding
There are many algorithms you can use to calculate the threshold without introducing user-bias. An evaluation of over 40 of these can be found in this paper:
Fiji has several plugins found in the menu Image › Adjust › Threshold for automatic calculation of an image threshold. These include Otsu’s thresholding, maximum entropy threshold, and mixture modelling thresholding. For a complete list of the methods available with Fiji see the Plugins section located in the Documentation section under the Content tab at the top of this page.
Watershed separation
Overlapping objects in a binary image can be separated using the menu command Process › Binary › Watershed.
First convert the image to binary by thresholding. The black pixels are then replaced with grey pixels of an intensity proportional to their distance from a white pixel. Black pixels closer to the edge are lighter than black pixels that are more central. This is the Euclidian distance map (EDM) of the black area. From this the centers of the objects are calculated. These are the ultimate eroded points (UEPs) of each black area meaning they are equidistant from each edge. These points are then dilated until they touch another black pixel. This meeting point is where a watershed line is drawn.
Analyze Particles
To analyze the particles in a segmented image, use the menu command Analyze › Analyze particles…. This will provide you with information about each particle in the image.
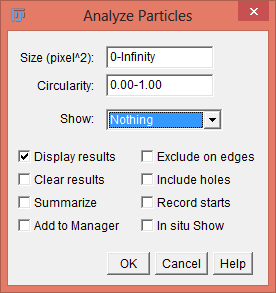
Set the minimum size and maximum pixel area size to exclude anything that is not an object of interest in the image. Roundness values between 0.0 and 1.0 can also be selected to help exclude unwanted objects. Select the Show: Outlines option to display an image of the detected objects. The Show drop-down menu also allows the user to show Nothing, Bare Outlines, Ellipses, Masks, Count Masks, Overlay Outlines, and Overlay Masks. The user can choose whether to Display results, Clear Results, Summarize, Add to Manager, Exclude on edges, Include holes, Record starts, and/or In situ Show.
The particle analysis can be automated via plugins or macros once the correct threshold value and particle size range has been determined for your objects of interest.
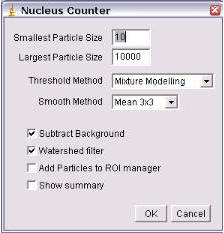
Nucleus Counter
This plugin automates many of the steps discussed above.
 .
.
- Enter the size range to be counted
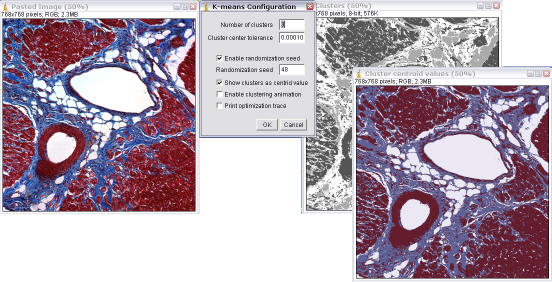
- Select the automatic thresholding method. This can be either Current, Otsu, Maximum Entropy, Mixture Modelling or k-means clustering. Current uses the threshold that has been set manually, see above.
- Perform a background correction.
- Use a Smooth filter.
- Perform a watershed separation.
- Add the particles to the ROI manager.
- Say yes to a summary.
Other options can easily be added on request.
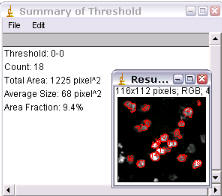
The count, area, and average size are returned as a text window and the outlined particles are overlaid on a duplicate of the original image.
Manual Counting
You can use the built-in Multi-point Tool to manually count particles.
Particle tracking
Particle Tracker Particle Tracker is a 2D feature point-tracking plugin for the automated detection and analysis of particle trajectories as recorded by video imaging in cell biology. The algorithm is decsribed in Sbalzarini and Koumoutsakos (2005[1]).
TrackMate Use the menu command Plugins › Tracking › TrackMate. This plugin allows you to perform single particle tracking of spot-like structures. For more in-depth information, see the TrackMate tutorial and explanation.
Manual Tracking Use the menu command Plugins › Tracking › Manual Tracking. This tool allows you to keep track of the movement of a cell.