This page was last revised for version 5.0.0.
SNT interacts with Labkit and Trainable Weka Segmentation (TWS), both leveraging machine learning algorithms for semantic segmentation of images (namely, random forest classifiers). The bridge between these tools makes it possible to:
- Import a pre-trained model into SNT and directly load the probability maps of the semantic segmentation as secondary tracing layer
- Train a model with SNT paths
The table below summarizes key differences between Labkit and TWS (as of SNT v4.3.0). Note that both tools classify images using the Weka framework.
| Labkit | TWS | |
|---|---|---|
| Image size | Out-of-core, multiple terabytes large image data | Smaller images (RAM-limited) |
| GPU support | via CLIJ2 | No |
| Underlying architecture(s) | ImgLib2 and BigDataViewer | ImageJ |
| Support for multichannel images | Yes | Yes (w/ caveats) |
| Support for timelapse images | Yes | Yes (w/ caveats) |
| Scripting and IJ macro language support | Yes. Some commands are macro-recordable | Yes. GUI is macro-recordable |
| Batch Processing | From GUI and via macros and scripts | Via macros and scripts |
| Import of pre-trained models into SNT | Yes | Yes |
| Direct loading of SNT paths as classification labels | Yes | Yes |
Importing Models
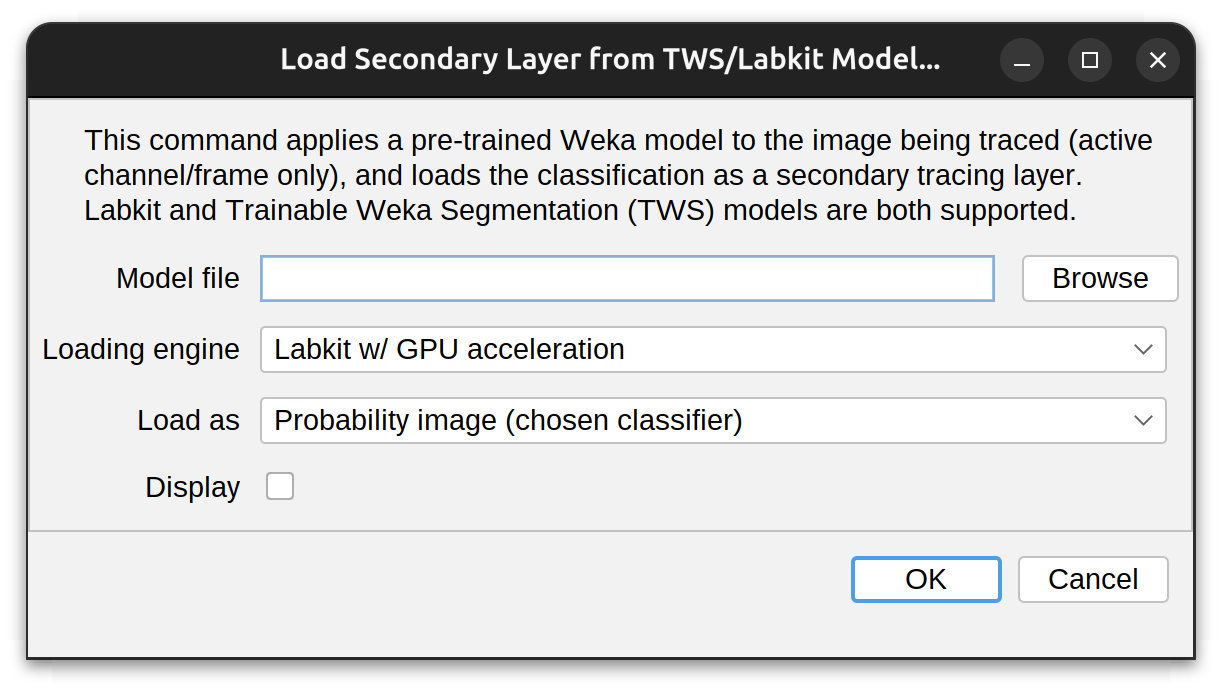
Importing of models can be done via the Secondary layer menu. Once the trained model is imported, it is applied to the image being traced, and the resulting classification is loaded as a secondary tracing image. The import prompt has the following options:
- Model file: The file to be imported. Typically, with a .model or .classifier extension (Labkit: JSON-encoded; TWS: XML-encoded)
- Loading engine: How the model should be loaded and applied. Either Labkit, Labkit w/ GPU acceleration or Trainable Weka Segmentation (TWS). NB: Labkit w/ GPU acceleration expects CLIJ2 access, and a CLIJ2-compatible graphics card
- Load as: Either Probability (p-map) or Segmented image. If Probability image, the class associated with neurite signal needs to be chosen in a follow-up prompt
- Display: Whether the classified image should be immediately displayed. NB: This can be done at anytime using the View› menu
Training Models
To convert traced paths into training labels, simply select the path(s) of interest and run the respective command in the Path Manager’s Process› menu. This will start up a new instance of Labkit/TWS preloaded with labels generated from selected path(s). Paths from different channels are split into distinct classification classes (i.e., 1 class per channel). Note that there are some (minor) idiosyncrasies in the way Labkit and TWS handle SNT-generated labels:
| Labkit | TWS | |
|---|---|---|
| Single-node paths | Valid labels | Typically skipped |
| Hyperstacks (images with CT dimensions) | Displayed by BigDataViewer | CT dimensions are displayed as a simple stack in the TWS window. IJ’s Stack to Hyperstack… command can be used to re-apply the original image layout to output images |
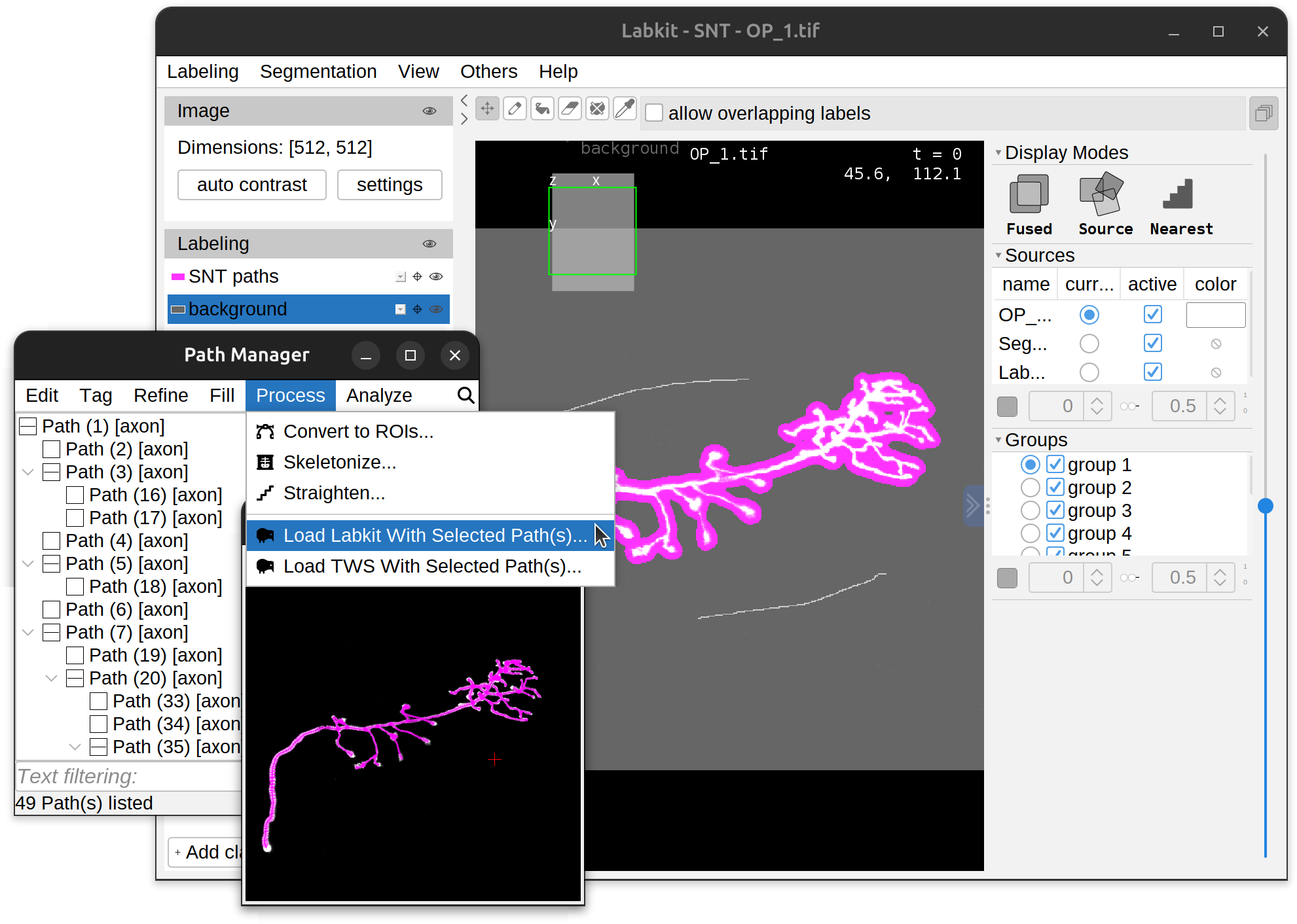
|
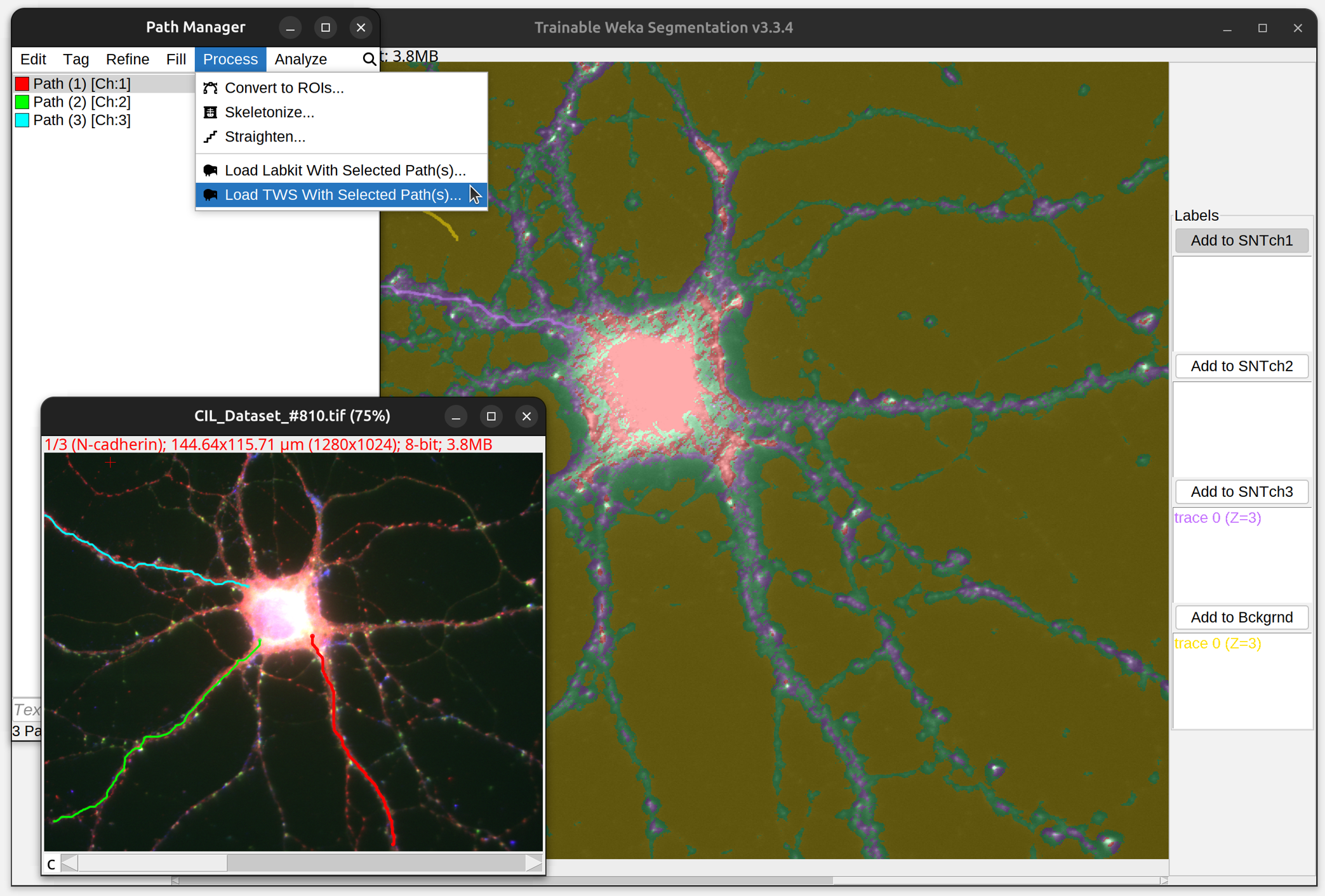
|
| Drosophila OP neuron (3D grayscale image, SNT's Demo 03) being classified in Labkit. | A triple-stained neuron (2D multichannel image, SNT's Demo 06) being classified in TWS. |
Scripts
There are a couple of examples in SNT’s neuroanatomy template collection handling image classification, namely:
- Apply Weka Model To Tracing Image: Demonstrates how to apply a pre-existing Weka model to the image being traced
- Train Weka Classifier: Exemplifies how to train a Weka model using traced paths and ROIs
References
The Weka framework is described in:
- Eibe Frank, Mark A. Hall, and Ian H. Witten (2016). The WEKA Workbench. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques”, Morgan Kaufmann, Fourth Edition, 2016. (PDF)
