The content of this page has not been vetted since shifting away from MediaWiki. If you’d like to help, check out the how to help guide!
When clicking Plugins › BigStitcher › BigStitcher in the the Fiji menu, it will open a macro-scriptable dialog that looks as following:
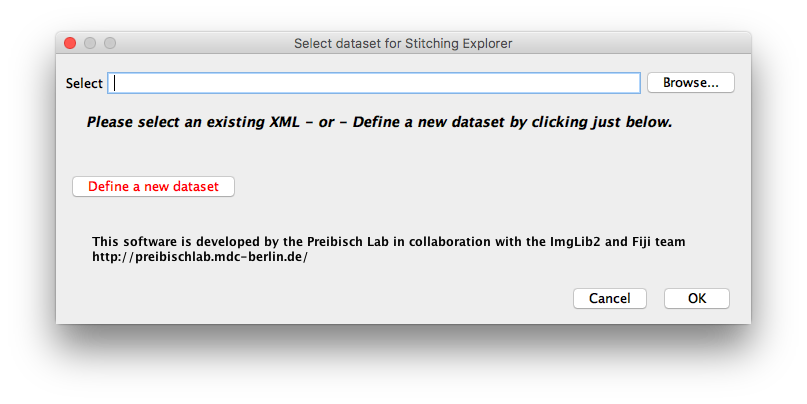
In order to define a new dataset, please click the respective button on the left side of the dialog. This will open a new window in which you can define how to import the image data. Since there is huge variety of data formats produced by various microscopy companies and self-built setups, we developed multiple types of importers:
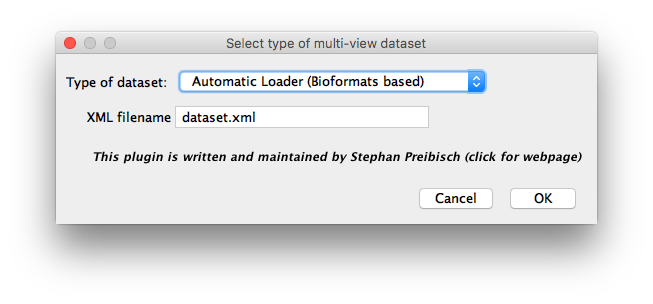
The Automatic Loader is a comprehensive importer that should work with most of image data. We highly suggest using this importer as a first choice, and just if you experience any problems to continue with one of the other options.
The Manual Loader (Bioformats) can be used to import datasets that consist of several stacks that can be opened using LOCI Bioformats. Manual assignment of the stacks to Angles, Tiles or Illuminations according to patterns in the filename is necessary.
The Manual Loader (TIFF only, ImageJ Opener)) can be used to import datasets that consist of several stacks that can be opened directly by ImageJ. Manual assignment of the stacks to Channels, Illuminations, Angles, Tiles or Illuminations according to patterns in the filename is necessary.
A single-purpose loader to import CZI-files produced by the Zeiss Lightsheet Z.1 microscope.
A single-purpose loader to import diSPIM datasets produced by MicroManager.
Note: Only one xml per directory is supported right now, even if they have a different filename.
Go back to the main page