This page was last revised for version 5.0.0.
The most up-to-date SNT API can be found at https://javadoc.scijava.org/SNT.
SNT Scripts
A key feature of SNT is its extensibility via scripts. This section provides an overview to the ever-growing list of SNT scripts. These can be accessed through the Plugins › NeuroAnatomy › Neuroanatomy Shortcut Window, the Scripts menu in the main dialog, or the Templates › Neuroanatomy menu in the Script Editor. Typically, holding ⇧ Shift while selecting a script from a menu outside the Script Editor will open it.
SNT Scripts are intentionally written in multiple languages to demonstrate flexibility, but the majority is written in Groovy and Python (Jython), for no other reason that those seem to be the most commonly used language by its developers. Script parameters are typically used for simpler routines.
Bundled Templates
Scripts menu in main dialog
There are 50+ scripts currently bundled in SNT. These are organized into non-rigid categories: Analysis (typically handling quantifications) , Batch (Bulk processing of files), Demos (teaching examples), Render (visualizations), Skeletons & ROIs (pixel-based analyses), Tracing (Tracing-related tasks), and Time-lapses (time-lapse utilities).
Bundled scripts can be accessed in two ways:
- Scripts menu in the main SNT dialog and SNT Shortcuts window (hold ⇧ Shift while selecting a script to have it open in the Script Editor)
- In the Templates › Neuroanatomy › list in the Script Editor
Your script can also be bundled with SNT, or made available through the Neuroanatomy update site. Did you write a useful routine? Please announce it, or submit a pull request so that it can be distributed with Fiji!
Adding Scripts to SNT
There are a couple of ways to have your own scripts appear in SNT’s Scripts menu:
Any script saved into Fiji’s “scripts” subdirectory containing SNT in the filename (e.g., C:\Users\user\Desktop\Fiji.app\scripts\My_SNT_Script.py or /home/user/Fiji.app/scripts/My_SNT_Script.py) will be listed in Scripts menu:
-
Go to Scripts › New… from the SNT dialog. You can choose to open an instance of Fiji’s Script Editor with preloaded boilerplate code in the scripting language of your choice or to assemble your boilerplate using the Script Recorder
-
Save the script in Fiji.app/scripts/ including ‘SNT’ anywhere in its file name
-
Run Scripts › Reload…, and your new script should appear in the full list of scripts found at Scripts › Full List….
Script Recorder
SNT features a script recorder that similarly to ImageJ’s macro recorder converts menu and button clicks into executable code. Note however that while the recorder captures simple commands well, it struggles to capture those that are more complex or particularly interactive.
The goal of the recorder is twofold: 1) simplify prototyping of new scripts and 2) Log your actions during a tracing session. This is particularly useful to assemble reproducible records.
There are two ways to start the recorder: Scripts › Record… or by pressing the Record button in Command Palette.
As a rule-of-thumb, commands that are simple or do not involve prompts record flawlessly. This includes setting filters for visibility of tags, applying Tags, or filtering paths in the Path Manager. Commands for fully automated reconstructions, or generating secondary layers should work well. However, many others remain limited.
REPL
SNT’s Scripting REPL (Read–Eval–Print Loop) is opened using Scripts › New › REPL. It is a Script Interpreter instance with pre-initialized variables that are entry-points to the API of current SNT session. The REPL serves as a commandline prompt with access to all of SNT classes.
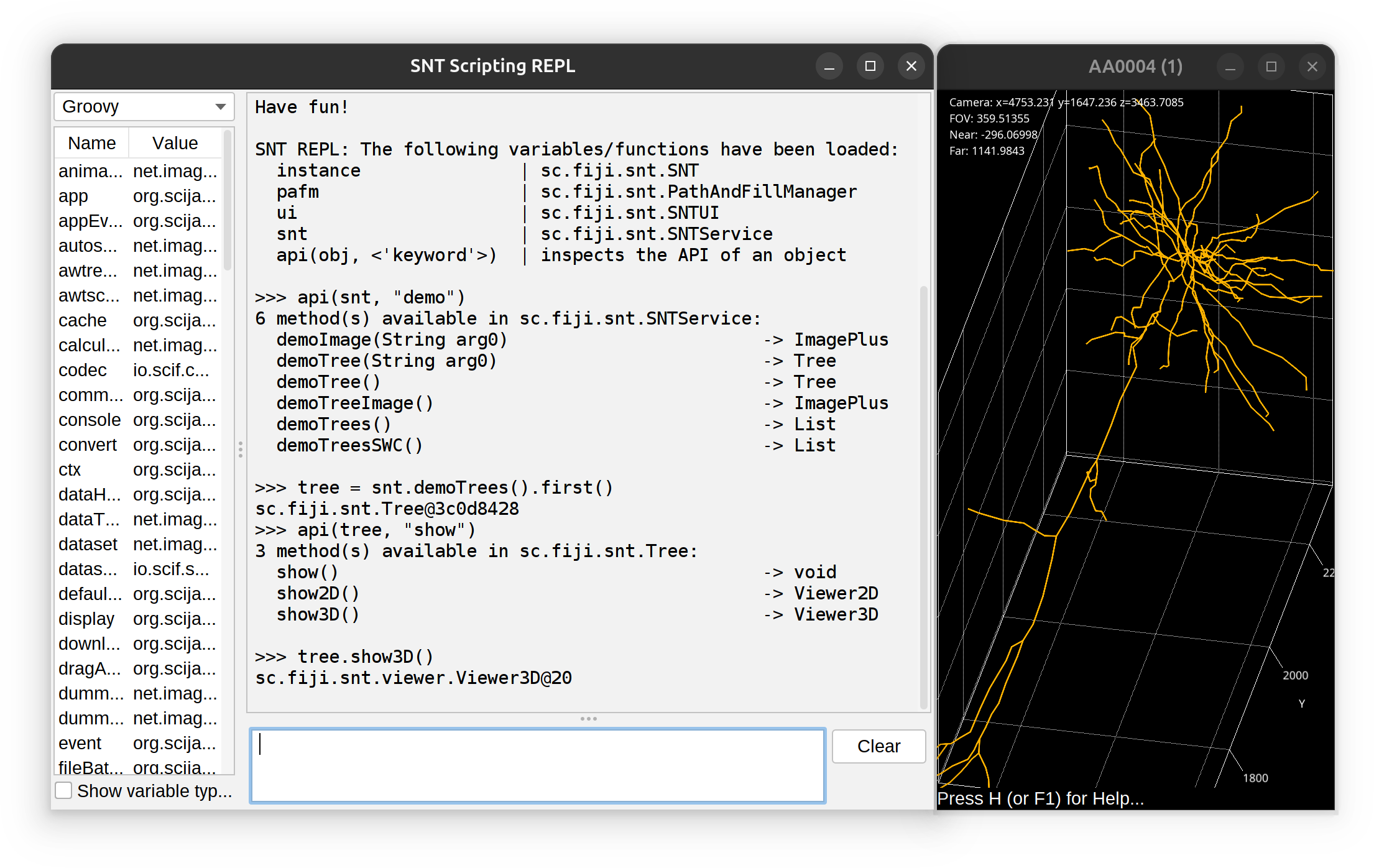
The REPL has access to all of SNT’s API. The prompt does not feature auto-completion but you can use api(object, 'optional keyword') to obtain a list of methods associated with an object. Example: To find out all of the ‘demo’ methods in SNTService, one would use:
>>> api(snt, "demo")
6 method(s) available in sc.fiji.snt.SNTService:
demoImage(String arg0) -> ImagePlus
demoTree(String arg0) -> Tree
demoTree() -> Tree
demoTreeImage() -> ImagePlus
demoTrees() -> List
demoTreesSWC() -> ListAnalysis of External Data
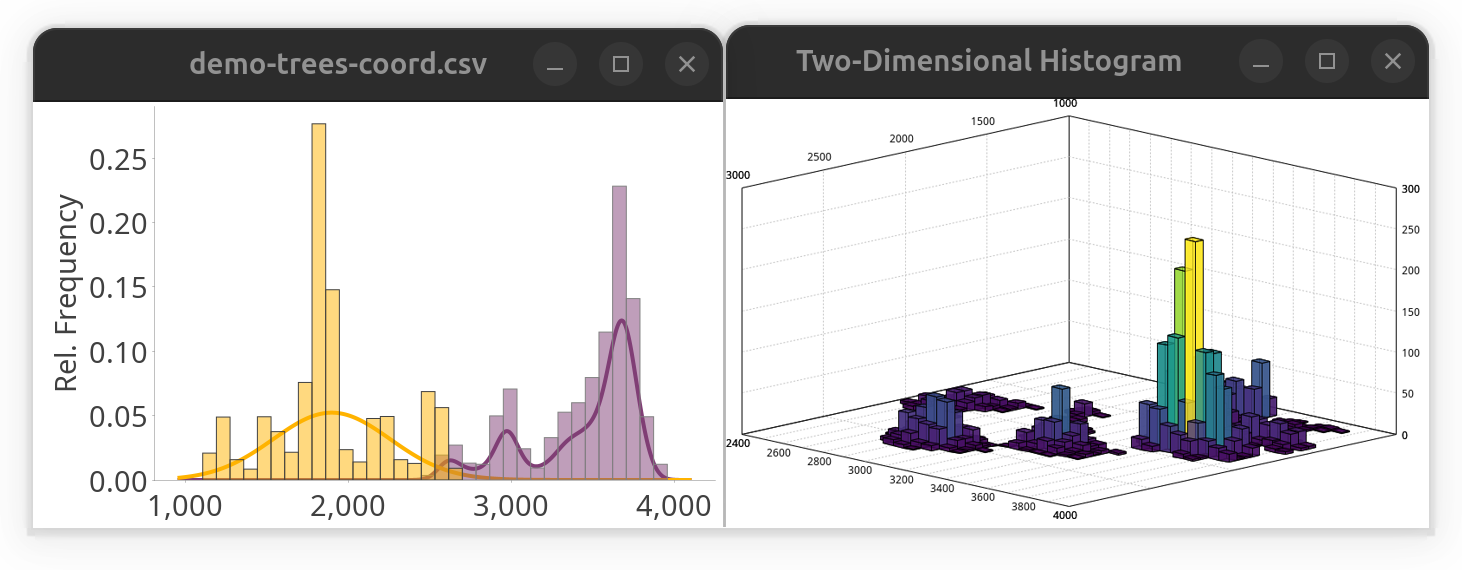
While SNT is not a statistical analysis software, it does offer some convenience methods to parse third-party data. E.g., to take a quick peek at a distribution of tabular values, one could use the following snippet to plot a histogram while fitting a Gaussian mixture model to the data. While it is written in Groovy, it would run almost verbatim in any other scripting language because it relies only on the SNT API.
import sc.fiji.snt.analysis.*
path = "https://raw.githubusercontent.com/morphonets/misc/master/dataset-demos/csv/demo-trees-coord.csv" // url or path to local file
table = SNTTable.fromFile(path) // read data into a table
headers = ["y", "z"] // headers of columns to be plotted
histogram = SNTChart.getHistogram(table, headers, false) // table, col. headers, radial plot?
histogram.setGMMFitVisible(true) // fit Gaussian mixed model to data
histogram.show() // display histogramAlternatively, the same data could be plotted in a two-dimensional histogram:
import sc.fiji.snt.analysis.*
import sc.fiji.snt.util.*
path = "https://raw.githubusercontent.com/morphonets/misc/master/dataset-demos/csv/demo-trees-coord.csv" // url or path to local file
table = SNTTable.fromFile(path) // read data into a table
SNTChart.showHistogram3D(table.get("y"), table.get("z"), ColorMaps.get("viridis")) // show 3D histogram of the same columns using viridis LUTNote that this approach would work for both local and remote files. The result of both snippets side-by-side:
Python Notebooks
Direct access to the SNT API from the Python programming language is made possible through the PyImageJ module. This enables full integration between SNT and any library in the Python ecosystem (numpy, scipy, etc.). The Notebooks directory in the SNT repository contains several examples at different complexity levels.
Here, we will only exemplify basic functionality. Please refer to the complete notebook examples for more details. Once the Python environment is properly setup, one can initialize Fiji:
import imagej
ij = imagej.init('sc.fiji:fiji', mode='interactive')Then, one import the needed SNT (Java) classes. E.g., to download a neuron reconstruction from the MouseLight database and calculate summary statistics on it, you would import the MouseLightLoader and TreeStatistics classes:
from scyjava import jimport
MouseLightLoader = jimport('sc.fiji.snt.io.MouseLightLoader')
TreeStatistics = jimport('sc.fiji.snt.analysis.TreeStatistics')Now you can access all the attributes and methods these classes offer. Let’s get a summary of the inter-node distances for a specific mouse cortical motor neuron (ID = “AA0100” in the MouseLight database).
def run():
loader = MouseLightLoader("AA0100")
if not loader.isDatabaseAvailable():
print("Could not connect to ML database", "Error")
return
if not loader.idExists():
print("Somehow the specified id was not found", "Error")
return
a_tree = loader.getTree('axon', None) # compartment, color
s_stats = TreeStatistics(a_tree)
metric = TreeStatistics.INTER_NODE_DISTANCE
summary_stats = s_stats.getSummaryStats(metric)
s_stats.getHistogram(metric).show()
print("The average inter-node distance is %d" % summary_stats.getMean())Fiji Scripting
Scripting in Fiji’s script editor is perhaps best done using Groovy and python. The latter as quite good autocompletion for objects that are not script parameters. The best way to start a new script is by choosing a boilerplate Scripts › New › From Template… in SNT or Neuroanatomy › Boilerplate in the script Editor. These templates hold boilerplate code in several programming languages (namely BeanShell, Groovy and Jython), and include the most essential imports and script parameters to facilitate rapid development.
Further Resources
As mentioned, SNT’s source code repository includes both Bundled Template Scripts and Jupyter notebooks. But additional snippets, examples, and further tutorials do exist online, including:
Example
Programmatic control over an open instance of Reconstruction Viewer (either called from within SNT or as a standalone application) can be achieved by selecting the Scripting › Record Script… command. It will then open an instance of SNT’s script recorder loaded with a boilerplate template containing the most essential imports and script parameters. The default programming language for this template can be chosen from the Recorder’s drop-down menu.
The following script exemplifies how to extend the boilerplate template to control the Viewer in real-time.
# Last tested: 2024-07-12, SNTv4.3.0pre
#@ Context context
#@ DatasetService ds
#@ DisplayService display
#@ ImageJ ij
#@ LogService log
#@ LUTService lut
#@ SNTService snt
#@ UIService ui
from sc.fiji.snt import Path, PathAndFillManager, SNT, SNTUI, Tree
from sc.fiji.snt.analysis import GroupedTreeStatistics, MultiTreeStatistics, NodeStatistics, TreeStatistics,\
ConvexHullAnalyzer, PersistenceAnalyzer, ShollAnalyzer, StrahlerAnalyzer, NodeColorMapper,\
TreeColorMapper, PathProfiler, PathStraightener, RoiConverter, SkeletonConverter, SNTChart, SNTTable
from sc.fiji.snt.analysis.graph import DirectedWeightedGraph
from sc.fiji.snt.analysis.sholl.parsers import TreeParser
from sc.fiji.snt.annotation import AllenCompartment, AllenUtils, VFBUtils, ZBAtlasUtils
from sc.fiji.snt.io import FlyCircuitLoader, MouseLightLoader, NeuroMorphoLoader
from sc.fiji.snt.plugin import SkeletonizerCmd, StrahlerCmd
from sc.fiji.snt.util import BoundingBox, PointInImage, SNTColor, SWCPoint
from sc.fiji.snt.viewer import Annotation3D, OBJMesh, MultiViewer2D, Viewer2D, Viewer3D
# Documentation Resources: https://imagej.net/plugins/snt/scripting
# Latest SNT API: https://javadoc.scijava.org/SNT/
# Rec. Viewer's API: https://javadoc.scijava.org/SNT/index.html?sc/fiji/snt/viewer/Viewer3D.html
# Tip: Scene details can be printed to Console using `viewer.logSceneControls()` or recorded
# by pressing 'L' (mnemonic: [L]og) when viewer is frontmost
# If opening this template from the standalone RV, the instance ID of the Viewer (e.g., 2)
# will be automatically passed as a parameter to getRecViewer()
viewer = snt.getRecViewer()
# Added code begins here ##################################################################
import time
def do_stuff(viewer):
viewer.logSceneControls()
# Load a neuron from Mouse Secondary Motor Area from the MouseLight database.
loader = MouseLightLoader("AA0100")
if not loader.isDatabaseAvailable():
ui.showDialog("Could not connect to ML database", "Error")
return
if not loader.idExists():
ui.showDialog("Somehow the specified id was not found", "Error")
return
# Extract nodes with tag "axon".
axon = loader.getTree('axon')
# Do the same for the dendrites.
dendrites = loader.getTree('dendrites')
# Load the Allen CCF Mouse Reference Brain and add it to the scene
viewer.loadRefBrain('mouse')
# Add both Trees to the scene
viewer.addTree(axon)
dendrites.setColor('cyan')
viewer.addTree(dendrites)
# Get the OBJMesh object representing the maximum depth Allen CCF compartment
# containing the soma point and add it to the scene.
soma_annotation = loader.getSomaCompartment()
viewer.addMesh(soma_annotation.getMesh())
# Enforce side perspective.
viewer.setViewMode('side')
# Next, we will applying a color mapping to the axon
# using branch order as the metric and the Ice Lut.
# Make an instance of TreeColorMapper using SciJava context.
mapper = TreeColorMapper(context)
# Get the net.imglib2.display.ColorTable object.
lut = mapper.getColorTable('Ice.lut')
# Color code the axon and render in scene.
# The min and max values returned by the mapper are used
# to set the range of the color bar legend.
bounds = viewer.colorCode(axon.getLabel(), TreeColorMapper.STRAHLER_NUMBER, lut)
viewer.addColorBarLegend(lut, bounds[0], bounds[1])
# Display a text annotation of the cell ID and metric used by the mapper.
scene_annotation = "{}: Branch Order".format(axon.getLabel())
viewer.addLabel(scene_annotation)
# Begin a rotation animation for the objects in scene.
viewer.setAnimationEnabled(True)
# Wait some time before applying a new color mapping
# based on path distances, using a different LUT (Fire).
time.sleep(5)
lut = mapper.getColorTable('Fire.lut')
# Remap the axon Tree and update scene.
bounds = viewer.colorCode(axon.getLabel(), TreeColorMapper.PATH_DISTANCE, lut)
viewer.addColorBarLegend(lut, bounds[0], bounds[1])
scene_annotation = "{}: Path Distance".format(axon.getLabel())
viewer.addLabel(scene_annotation)
do_stuff(viewer)