Introduction
This last part on particle-linking modules concludes the series of tutorials on extending TrackMate. The most difficult modules to create are spot detectors, which was the subject of the previous tutorial. Particle-linking modules, or trackers, are a little bit less complicated.
However, you still need to understand how we store and manipulate links in TrackMate, and this implies very briefly introducing mathematical graphs.
Simple, undirected graphs
TrackMate stores the results of the detection step as spots in a SpotCollection. The tracking results are mainly links between these spots so we needed a structure to hold them. We went for the most general one, and picked a mathematical graph.
Mathematical graphs are mathematical structures that hold objects (vertices) and links between them (edges, we will use the two terms interchangeably). TrackMate relies specifically on a specialization: it uses an undirected, simple weighted graph.
- Undirected means that a link between A and B is the same as a link between B and A. There is no specific direction and it cannot be exploited. However, you will see that the API offers specific tools that can fake a direction. Indeed, since we deal mainly with time-lapse data, we would like to make it possible to say that we iterate a graph following the time direction.
- Simple is not related to the efforts that must be made to grasp this mathematical field, but to the fact that there can be only 1 or no link between two spots, and that we do not authorize a link going from one spots to this same spot (no loop).
- Weighted means that each link has a numerical value, called weight, associated to it. We use it just to store some of the results of the tracking algorithm, but it has no real impact on TrackMate.
This restrictions do not harm the generality of what you can represent in Life Science with this. You can still have the classical links you find in typical time-lapse experiment:
- Following a single object over time:
A0 - A1 - A2 - A3 - ...
- A cell division:
A0 - A1 -+- B2 - B3 - ...
|
+- C2 - C3 - ...
- But also anything fusions, tripolar divisions, and a mix of everything in the same model.
Graphs in TrackMate
On a side note, this is important if you plan to build analysis tools for TrackMate results. TrackMate philosophy is to offer managing the most general case (when it comes to linking), but your analysis tools might require special use cases.
- For instance, when you are tracking vesicles that do not fuse nor split, you just have a linear data structure (an array of objects for each particle).
- When you follow a cell lineage, you have a rooted mathematical tree.
- And if all cells divide in two daughters, then you have a rooted binary tree.
They all are specialization of the simple graph, and offer special tools that can be very useful. But TrackMate assumes none of these specializations. It stores and manipulate a graph.
Since we are Java coders, we use a Java library to manipulate these graphs, and for this we rely on the excellent JGraphT library. Why a graph? Why not storing a list of successors and a list of predecessors for each spot? Or directly have a track object that would save some time on determining what are the tracks? Well, because a graph is very handy and simple to use when creating links. When you will write your own tracker, and found a link you want to add the model, the only thing you have to do is: graph.addEdge(A, B). You don’t have to care whether A belongs to a track and if yes to what track, you don’t need to see the whole graph globally, you can just focus on the local link. Adding a link in the code is always very simple.
Then of course, you still need a way to know how many tracks are there in the model, and what are they made of. But this is the job of TrackMate. It offers the API that hides the graph and deals in track. This is done via a component of the model, the TrackModel. But in the tracker we will not use this. We will be given a simple graph, and will have to flesh it out with spots and links between these spots. When the tracker completes, TrackMate will build and maintains a list of tracks from it.
The price to pay for this simplicity is that - when tracking - it is not trivial to get the global information. For instance, it is easy to query whether a link exists between two spots, but the graph does not see the tracks directly. If you need them, you either have to build them from the graph, either have to maintain them locally. But more on this below.
Particle-linking algorithms in TrackMate
We used the term tracker since the beginning of this series, but the correct term for what we will build now is particle linking algorithm. Our particles are the visible spots resulting from the detection step, and the links will be the edges of the graph. A tracker could be defined as the full application that combines a particle detection algorithm with a particle linking algorithm.
In TrackMate, particle linking algorithms implements the SpotTracker.java interface. It is very simple. As explained in the docs, a SpotTracker algorithm is simply expected to create a new SimpleWeightedGraph from the SpotCollection given (using of course only the visible spots). We use a simple weighted graph:
- Though the weights themselves are not used for subsequent steps, it is suggested to use edge weight to report the cost of a link.
- The graph is undirected, however, some link direction can be retrieved later on using the
Spot.FRAMEfeature. The SpotTracker implementation does not have to deal with this; only undirected edges are created. - Several links between two spots are not permitted.
- A link with the same spot for source and target is not allowed.
- A link with the source spot and the target spot in the same frame is not allowed. This must be enforced by implementations.
There is also an extra method to pass a instance of Logger to log the tracking process progresses. That’s all.
A dummy example: drunken cell divisions
There is already an example online that does random linking. Let’s do something else, and build a tracker that links a spot to any two spots in the next frame (if they exist) as if it would go cell division as fast as it can.
Creating the class yields the following skeleton:
package plugin.trackmate.examples.tracker;
import org.jgrapht.graph.DefaultWeightedEdge;
import org.jgrapht.graph.SimpleWeightedGraph;
import fiji.plugin.trackmate.Logger;
import fiji.plugin.trackmate.Spot;
import fiji.plugin.trackmate.tracking.SpotTracker;
public class DrunkenCellDivisionTracker implements SpotTracker
{
private SimpleWeightedGraph< Spot, DefaultWeightedEdge > graph;
private String errorMessage;
private Logger logger = Logger.VOID_LOGGER;
@Override
public SimpleWeightedGraph< Spot, DefaultWeightedEdge > getResult()
{
return graph;
}
@Override
public boolean checkInput()
{
return true;
}
@Override
public boolean process()
{
graph = new SimpleWeightedGraph< Spot, DefaultWeightedEdge >( DefaultWeightedEdge.class );
return true;
}
@Override
public String getErrorMessage()
{
return errorMessage;
}
@Override
public void setNumThreads()
{
// Ignored. We do not multithreading here.
}
@Override
public void setNumThreads( final int numThreads )
{
// Ignored.
}
@Override
public int getNumThreads()
{
return 1;
}
@Override
public void setLogger( final Logger logger )
{
// Just store the instance for later use.
this.logger = logger;
}
}
Parameters need to be passed to the class via its constructor. As for detectors, the factory we will build later will be in charge of getting these parameters. Of course, the most important one is the SpotCollection to track. In our case it will be the only one, as our dummy tracker do not have any settings. So we can have a constructor like this:
public DrunkenCellDivisionTracker( final SpotCollection spots )
{
this.spots = spots;
}
then we exploit the SpotCollection in the process() method. Our strategy here is to loop over all the frames that have a content, and link each spot to two spots in the next frame - wherever they are - until there is either no source spots or no target spots left. The method looks like this:
@Override
public boolean process()
{
graph = new SimpleWeightedGraph< Spot, DefaultWeightedEdge >( DefaultWeightedEdge.class );
// Get the frames in order.
final NavigableSet< Integer > frames = spots.keySet();
final Iterator< Integer > frameIterator = frames.iterator();
// Get all the visible spots in the first frame, and put them in a new
// collection.
final Iterable< Spot > iterable = spots.iterable( frameIterator.next(), true );
final Collection< Spot > sourceSpots = new ArrayList< Spot >();
for ( final Spot spot : iterable )
{
sourceSpots.add( spot );
}
// Loop over frames, and link the source spots to spots in the next
// frame.
double progress = 0;
while ( frameIterator.hasNext() )
{
final Integer frame = frameIterator.next();
final Iterator< Spot > it = spots.iterator( frame, true );
SOURCE_LOOP: for ( final Spot source : sourceSpots )
{
/*
* Add the source to the graph, if it is not already done (doing
* it several time is not a problem: it's backed up by a Set).
*/
graph.addVertex( source );
// Finds 2 targets.
for ( int i = 0; i < 2; i++ )
{
if ( it.hasNext() )
{
final Spot target = it.next();
// You must add it as vertex before creating the link.
graph.addVertex( target );
// This is how we create a link.
final DefaultWeightedEdge edge = graph.addEdge( source, target );
// We get the edge back, and set its weight through:
if ( null != edge )
{
graph.setEdgeWeight( edge, 3.14 );
/*
* Edge can be null if a link already exists between
* the two spots.
*/
}
}
else
{
break SOURCE_LOOP;
}
}
}
// Regenerate source list for next frame.
sourceSpots.clear();
for ( final Spot spot : spots.iterable( frame, true ) )
{
sourceSpots.add( spot );
}
progress += 1;
logger.setProgress( progress / frames.size() );
}
return true;
}
So it’s not really complicated. Which is good, because the complicated part, completely omitted here, is the one where you have to determine what links to create. This is where you Science should kick in.
The factory class
Now that we have the clever part of the code (the one that does the actual linking), we need to deal with TrackMate integration. Like for the detection modules, this is done via a factory class, named SpotTrackerFactory.java. It is completely equivalent to the SpotDetectorFactory we saw in the previous tutorial, so I won’t detail the common methods again.
The methods specific to the tracker are:
public SpotTracker create( final SpotCollection spots, final Map< String, Object > settings );
This method instantiates the actual tracker class. You can see that it received the SpotCollection and a settings map. This method is expected to unpack this map and extract the actual parameters need to instantiate the tracker.
Note that contrary to the detector factory, TrackMate calls this method only once for a tracking process. It does not generate a tracker per frame. So it is actually simpler than for detection: a tracker instance is expected to solve the tracking problem for the whole model at once. Therefore, there is no need for a setTarget() method, like previously.
public ConfigurationPanel getTrackerConfigurationPanel( final Model model );
This method should generate a GUI panel to request tracking parameters from the user. Completely similar to the detection modules.
marshallandunmarshall
Save to and retrieve from XML, like previously.
public String toString( final Map< String, Object > sm );
Used to pretty-print the settings map specific to this tracker.
The rest is classic. Here is what it looks like for our tracker:
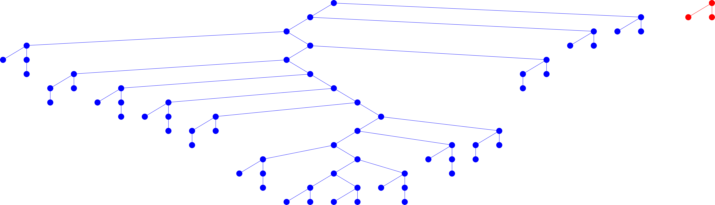
TrackMate recognize there were two tracks. You did not have to worry about that.
Controlling the visibility of your tracker with the SciJava visible parameter
Our tracker is a good dummy example, but it is not that useful. There might be cases where you wish for a TrackMate module not to be visible in the GUI. There is way to do that, just by tuning the SciJava annotation:
To make a TrackMate module available in TrackMate, but not visible in the GUI menus, use the annotation parameter visible = false
So editing the header of our tracker factory to make it look like:
@Plugin( type = SpotTrackerFactory.class, visible = false )
public class DrunkenCellDivisionTrackerFactory implements SpotTrackerFactory
is enough to hide it in the menu. This is different from the enabled parameter we saw in one the previous tutorial. The factory is instantiated and available in TrackMate; it just does not show up in the menu.
Wrapping up
The full code, as well as the code for another tracker example can be found on GitHub: Tracker code. And this concludes flatly our series of tutorials on how to extend TrackMate. Go forth now, and bend it to your needs; it is your tool.
Jean-Yves Tinevez 09:26, 5 September 2014 (CDT)