The advanced Omnipose integration in TrackMate works roughly as the Omnipose integration. The omnipose advanced detector gives you the possibility to tune some Omnipose model hyper-parameters. It requires Omnipose to be installed on your system and working independently. This page gives installation details and advices at how to use the omnipose advanced integration in TrackMate.
Omnipose
Omnipose is a segmentation algorithm based on Deep-Learning techniques, and inspired from the Cellpose architecture. Omnipose is well suited for bacterial cell segmentation, and achieves remarkable performances on mixed bacterial cultures, antibiotic-treated cells and cells of elongated or branched morphology.
If you use the Omnipose TrackMate module for your research, please don’t forget to cite the Omnipose paper:
doi:10.1038/s41592-022-01639-4
Example
E. coli, Marie Anselmet and Rodrigo Arias Cartin, Barras lab, Institut Pasteur
Installation
TrackMate-Cellpose update site
The TrackMate-Omnipose-Advanced module is part of the TrackMate-Cellpose Fiji extension. Please check the installation instruction on the TrackMate-Cellpose page.
Omnipose installation
See the installation instructions on the basic Omnipose integration. page.
The integration works with Omnipose version 1.0.6.
Like for the cellpose integration, you need to have a working Python installation of omnipose on the computer you want to use this extension with. To install Omnipose, you can refer directly to the installation guide provided on the Omnipose release page on PypI, but some steps may generate errors.
We give below a tested procude to install omnipose on a Mac Intel, Mac M1 to M3 and Windows with GPU support via mamba.
An example Windows installation working on GPU (may need to adapt the cuda version to your drivers):
mamba create -n omnipose-106 'python==3.9.18' pytorch torchvision pytorch-cuda=11.8 -c pytorch -c nvidia -y
mamba activate omnipose-106
pip install natsort
pip install scipy==1.11.4
pip install omnipose==1.0.6
An example Mac Intel installation:
mamba create -n omnipose-106 'python==3.9.18'
mamba activate omnipose-106
mamba install pytorch torchvision -c pytorch
pip install natsort
pip install scipy==1.11.4
pip install fastremap==1.12.2
pip install omnipose==1.0.6
An example Mac M1 to M3 installation:
mamba create -n omnipose-106 'python==3.9.18'
mamba activate omnipose-106
mamba install pytorch torchvision -c pytorch
pip install natsort
pip install scipy==1.11.4
pip install omnipose==1.0.6
Omnipose advanced hyper-parameters
Choosing the Omnipose advanced detector gives you the possibility to tune the values of the mask and the flow threshold, which are fixed to their default values in the Omnipose detector.
The flow_threshold parameter is the maximum allowed error of the flow fields averaged over all pixels in a given mask. The default is flow_threshold=0.4.
We recommend you to increase this threshold if Omnipose is not returning as many masks as you expect, and to decrease it if Omnipose is returning too many spurious masks.
The mask threshold is applied to the distance transform output of Omnipose to seed cell masks pixels. The default is mask_threshold=0.0. You should decrease this threshold if Omnipose is not returning as many masks as you expect, or if the returned masks do not cover the entire cell.
More details about Omnipose hyper-parameters can be found here.
Troubleshooting “Found 0 spots” errors with pretrained models
On some systems we have noticed that sometimes TrackMate returns 0 detections for the cellpose and omnipose detectors, even when the installation of these two programs worked correctly. In most cases, this is due to the fact that the pretrained models have not been downloaded prior to running the TrackMate integration. If the model your trying to call doesn’t exist on your computer, a message should appear in the TrackMate logger after the preview step. To fix this, the easiest way is to launch the omnipose Python GUI, and segment a single small image. This will trigger the download of the pretrained models. After this, the TrackMate Omnipose integration should work as expected.
Custom models
You can use in this TrackMate-Omnipose-Advanced integration your own custom models that were trained on the same version 1.0.6 of Omnipose. You should select the option Custom in the list of models, and provide the path of the custom model you want to use.
In the last versions of Omnipose, some changes were made, in particular in the way to call the models and the default parameters values. We made our code as robust as possible to deal with these changes but we can’t ensure this TrackMate integration of Omnipose is compatible with custom models trained on all the previous versions of Omnipose.
Tutorial
The following tutorial is identical to the one on the basic Omnipose integration page, but displays the changes you can find in the advanced version. We segment and track individual bacteria in several colonies. The source video follows E. coli bacteria imaged in phase contrast. We want to segment them, track them, and plot the growth curve for the bacteria imaged.
The image file can be found on Zenodo:
Download it and save it somewhere convenient.
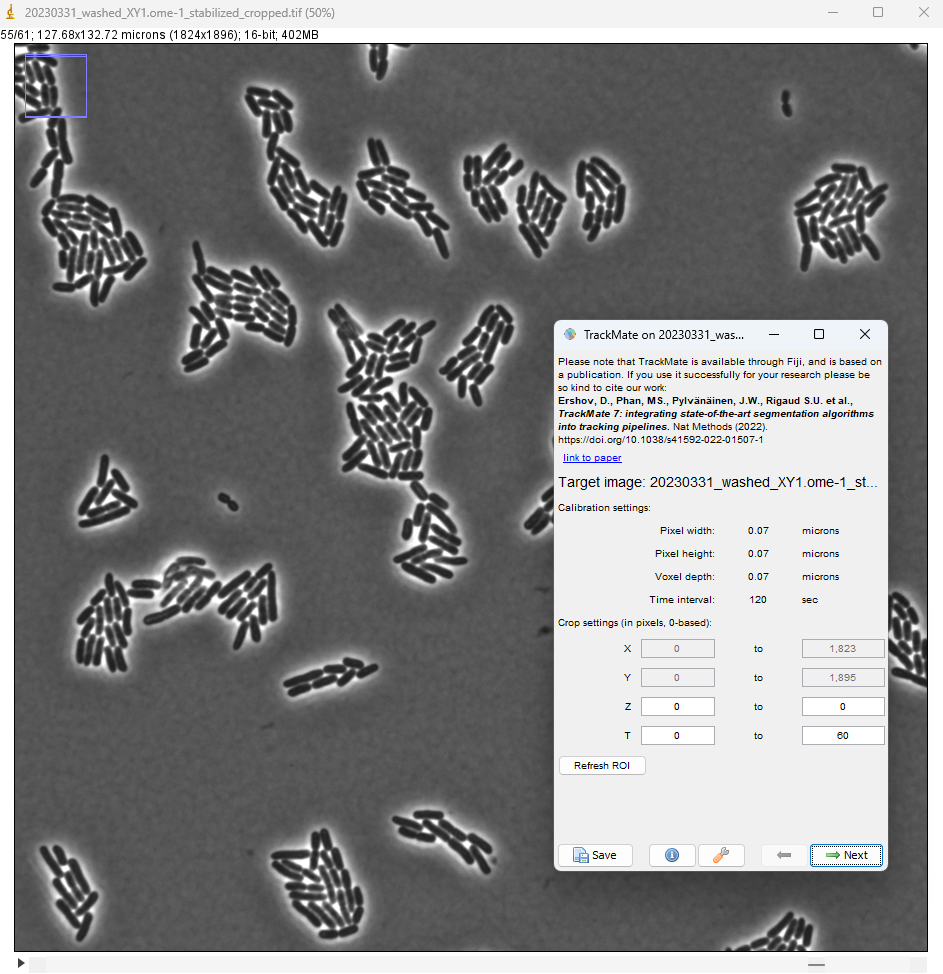
After executing the installation procedure above, launch Fiji and open the image. It is a relatively large image with a small pixel size (70 nm). Each 2D frame is 1824 x 1896 and a bacteria of length 4 um is imaged over about 50 pixels. With this image selected, run TrackMate (Plugins > TrackMate).
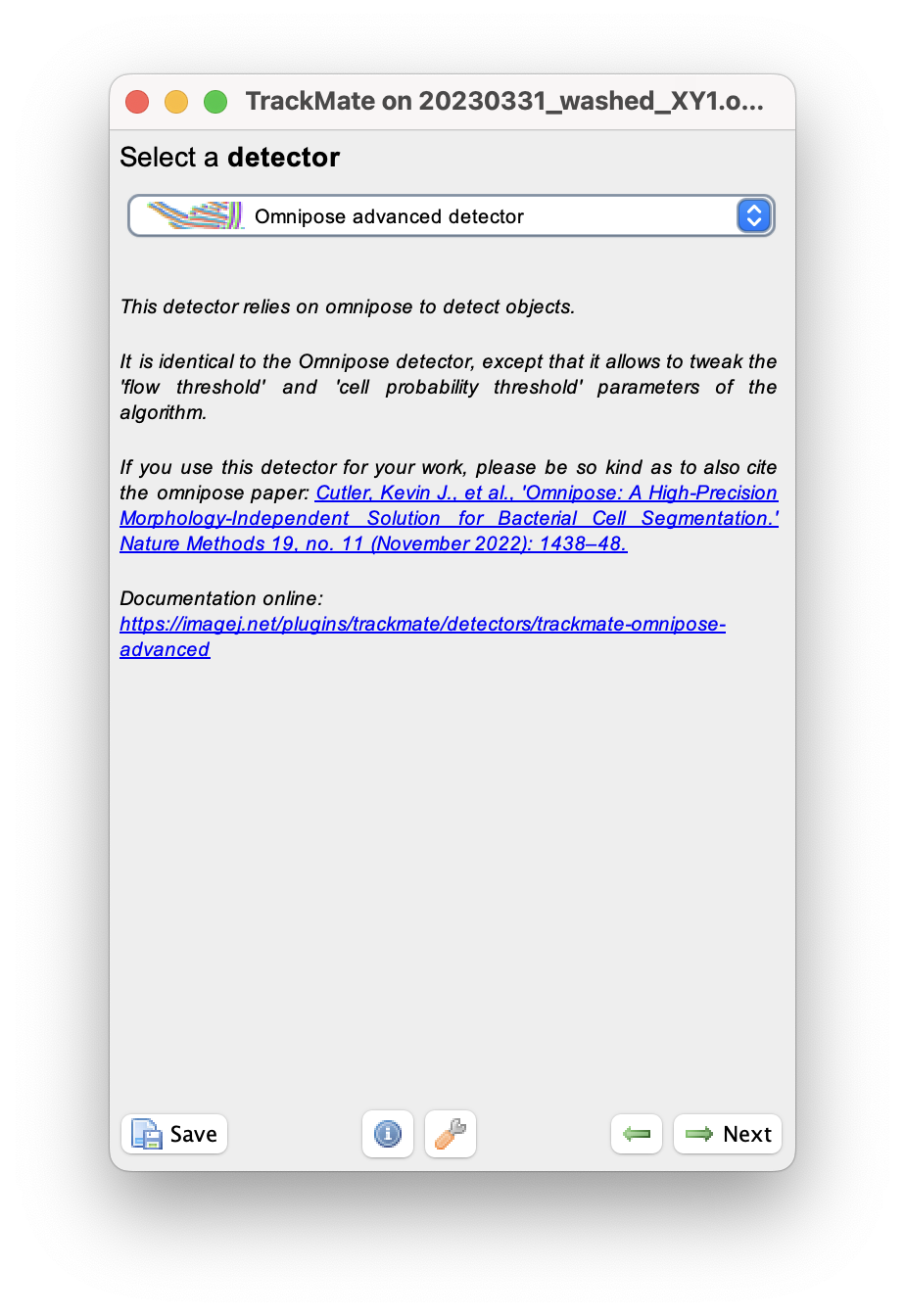
In the detector selection panel, pick the Omnipose advanced detector.
The configuration panel is quasi identical to that of the omnipose detector. Here, the default values will give us satisfactory results. You just need to set the conda environment to be the one where you installed Omnipose.
The values of the flow and mask thresholds are set with two sliders and number inputs.
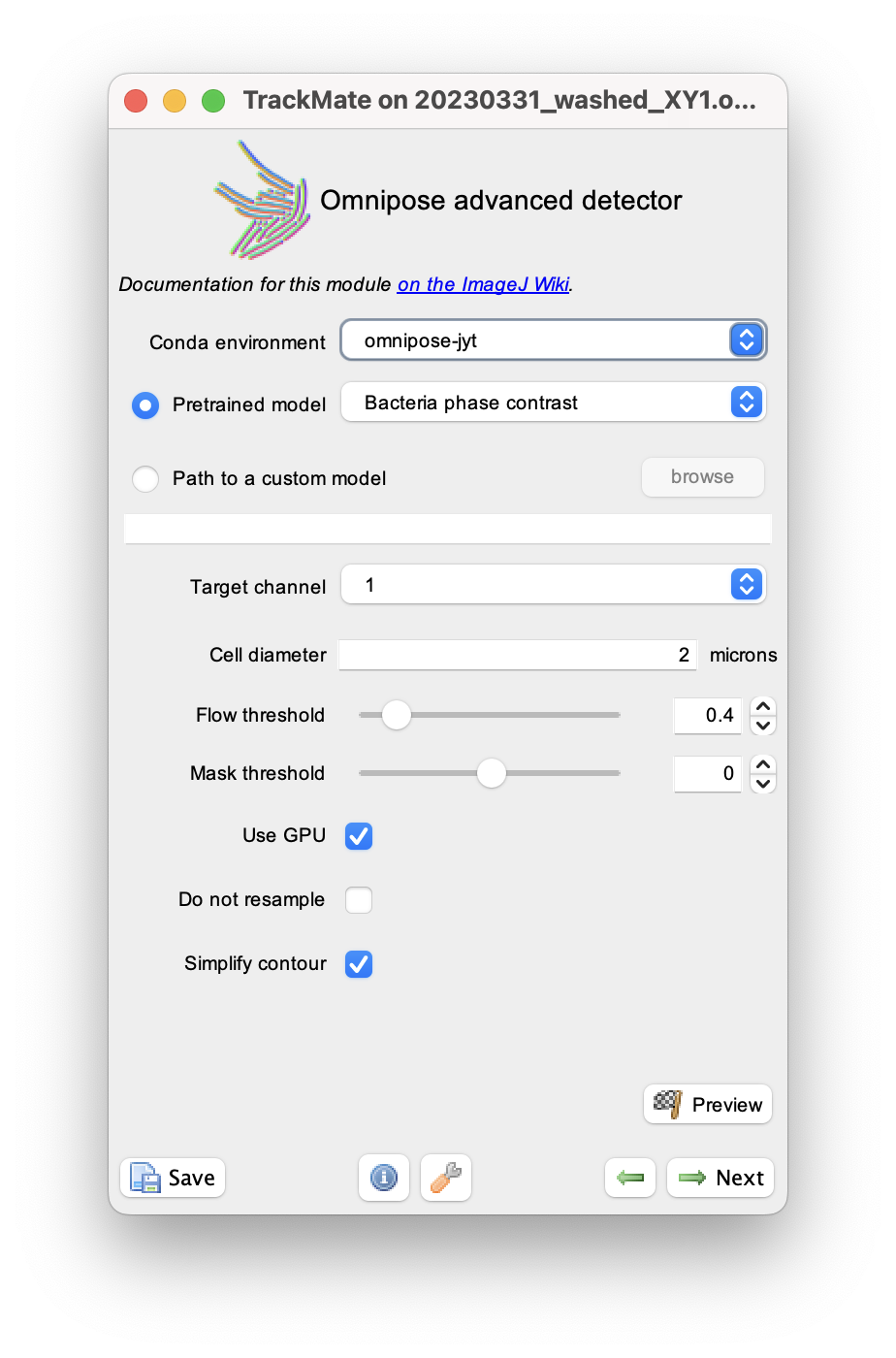
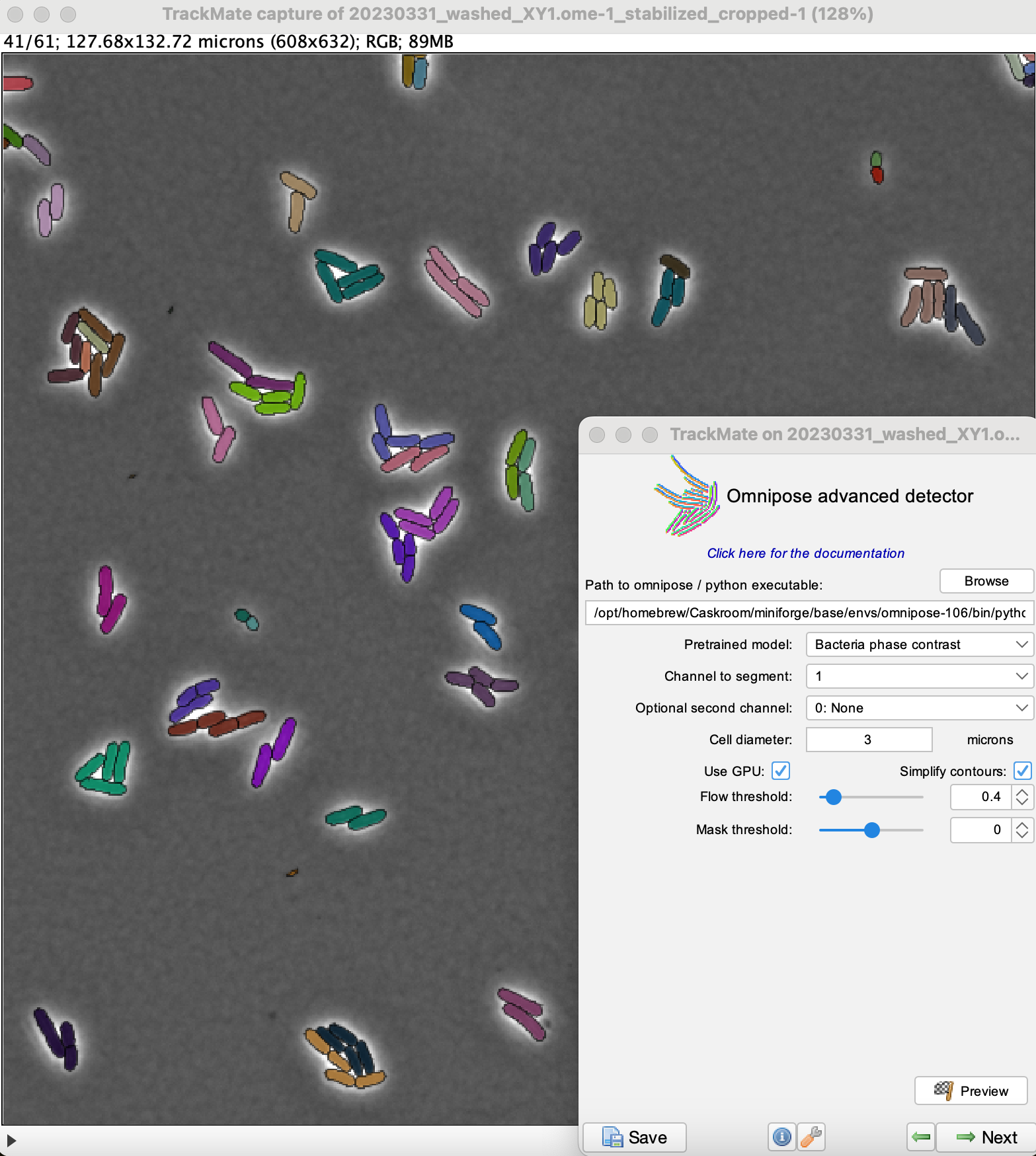
Click the Preview button to check that the settings give correct results.
Here the quality is equal to the number of pixels inside detected objects.
We see that there are some very small objects with settings we have, but otherwise the results are excellent with the default parameters.
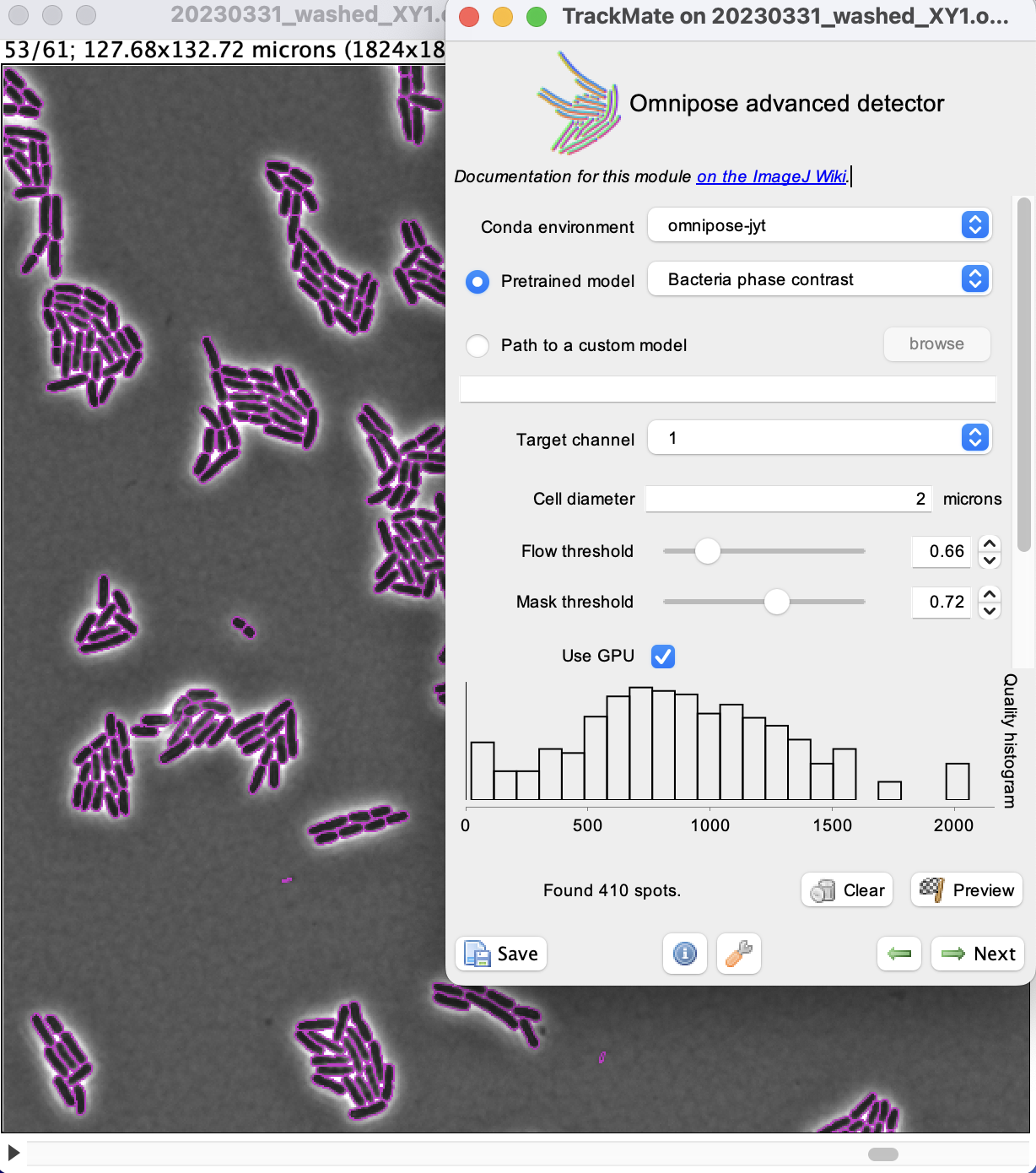
We can now run the detection over the whole movie by clicking the Next button.
On my windows machine with a NVIDIA 2080Ti, it takes about 3 to 4 seconds per frame.
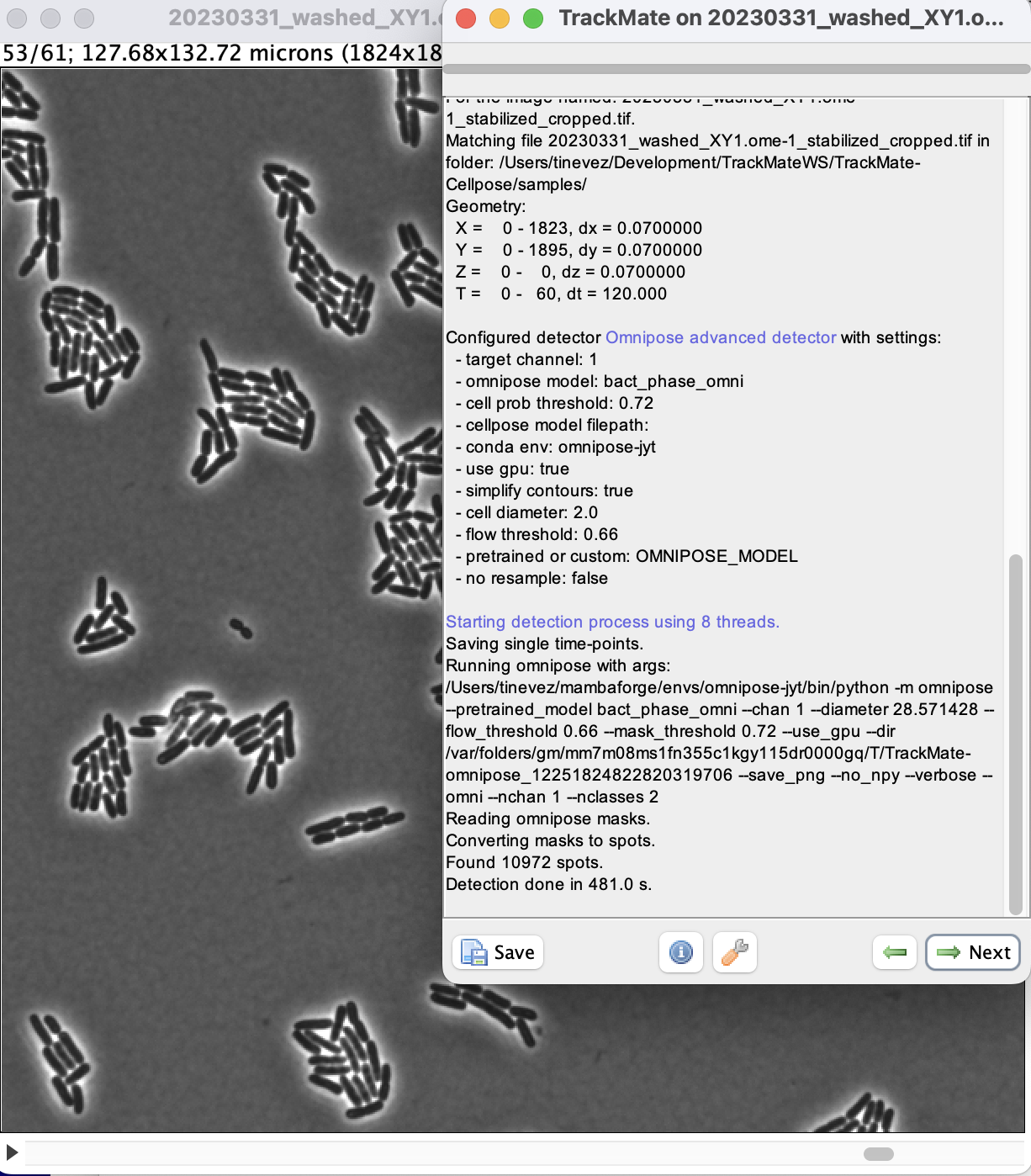
When done, click Next to reach the initial filtering panel.
The quality histogram displays a small peak at low quality, corresponding to small spurious objects.
We can filter them out by setting the threshold in between the two peaks in the histogram.
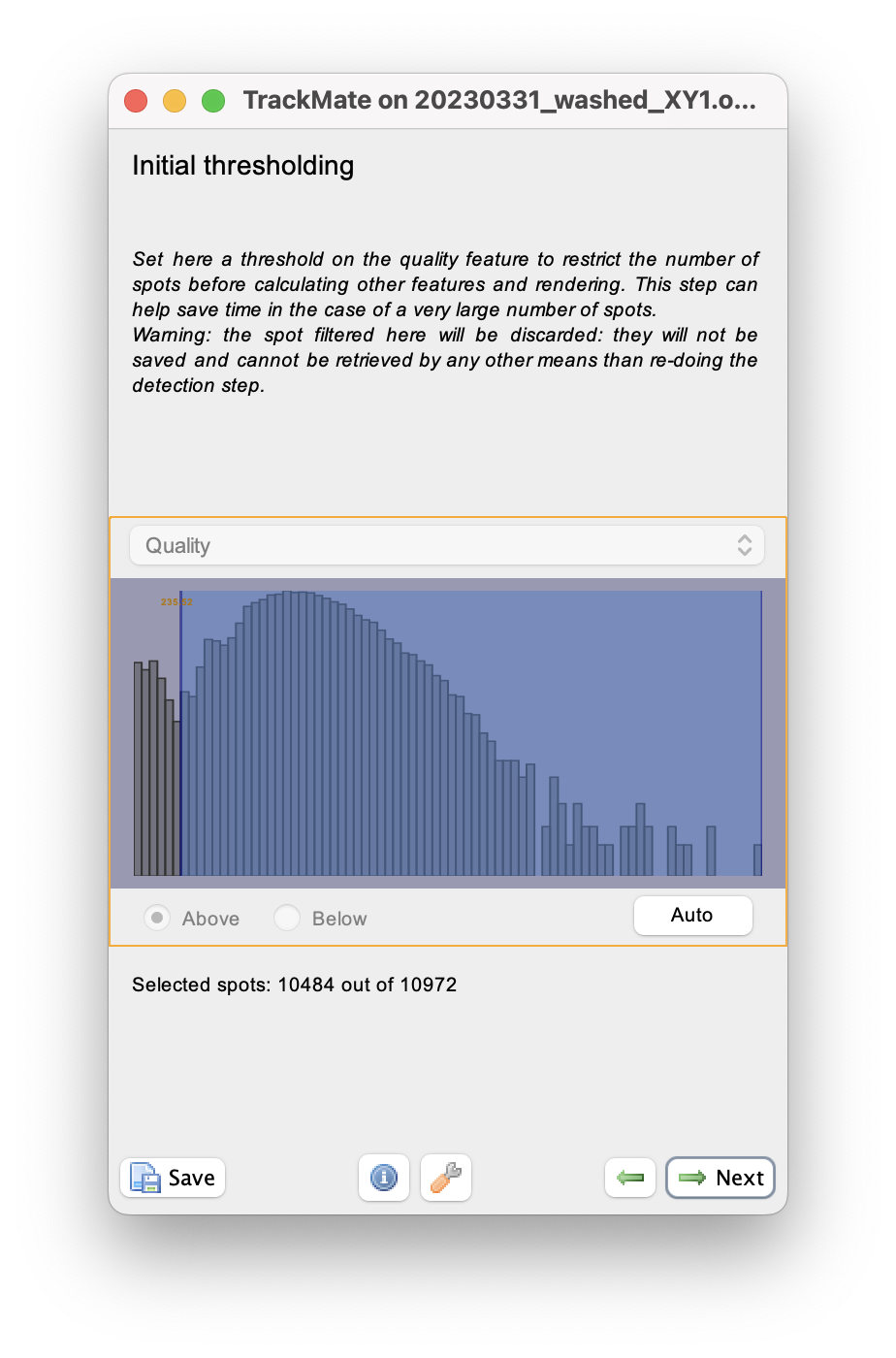
We can skip filtering objects.
Click Next until you reach the tracker selection panel.
The bacteria do not move much, but divide, pushing each other away.
We found out that for this kind of dynamics the Overlap tracker gives good tracking results.

A few parameter needs to be tuned.
For the Min IoU we use 0.1.
For the Scale factor we use 1.3.
Click Next.

This yields a good lineage of the bacteria. We see each “colony” grows, and mix with the close ones. With the tracker settings we picked above, the lineage of each mother bacteria is preserved rather well, even when a colony touches its neighbors.
To color each bacteria by its colony, on the Display options panel click on the Edit settings button.
In the display settings window that appear, make the following changes:
- select
draw spots filled - set the
spot alpha transparencyto 0.7 - for
spot color, select Track index - uncheck
draw tracks
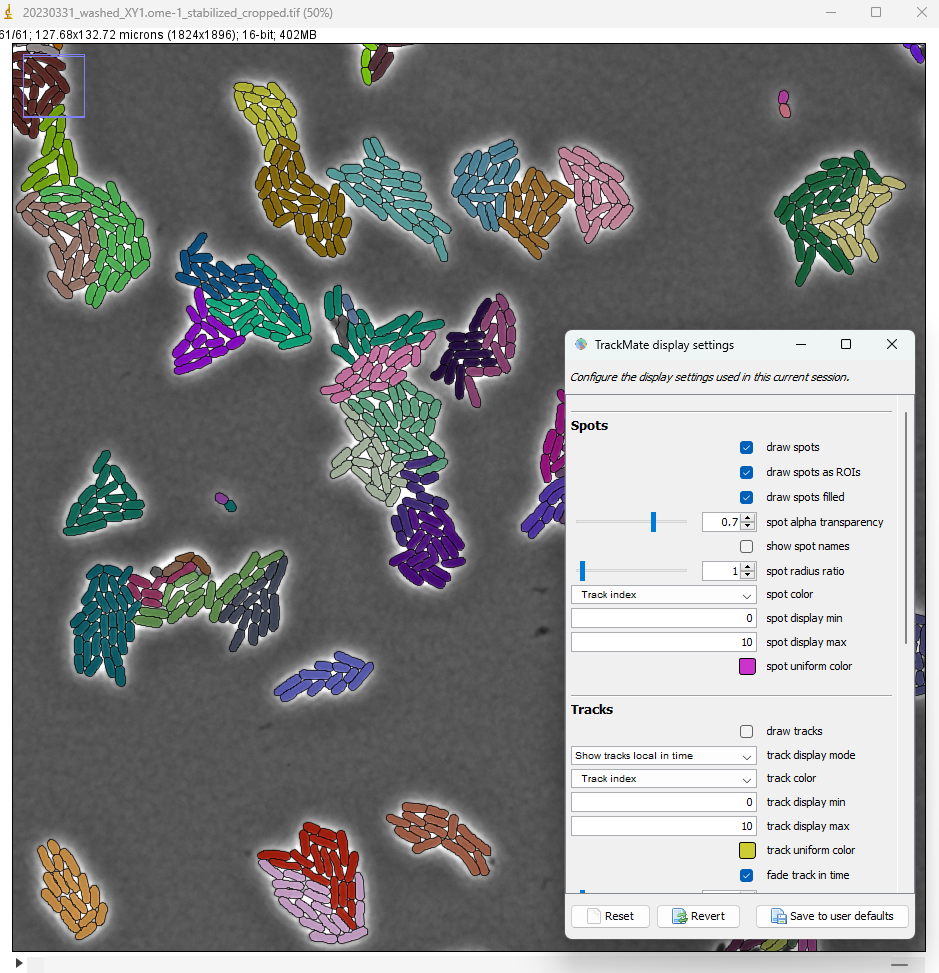
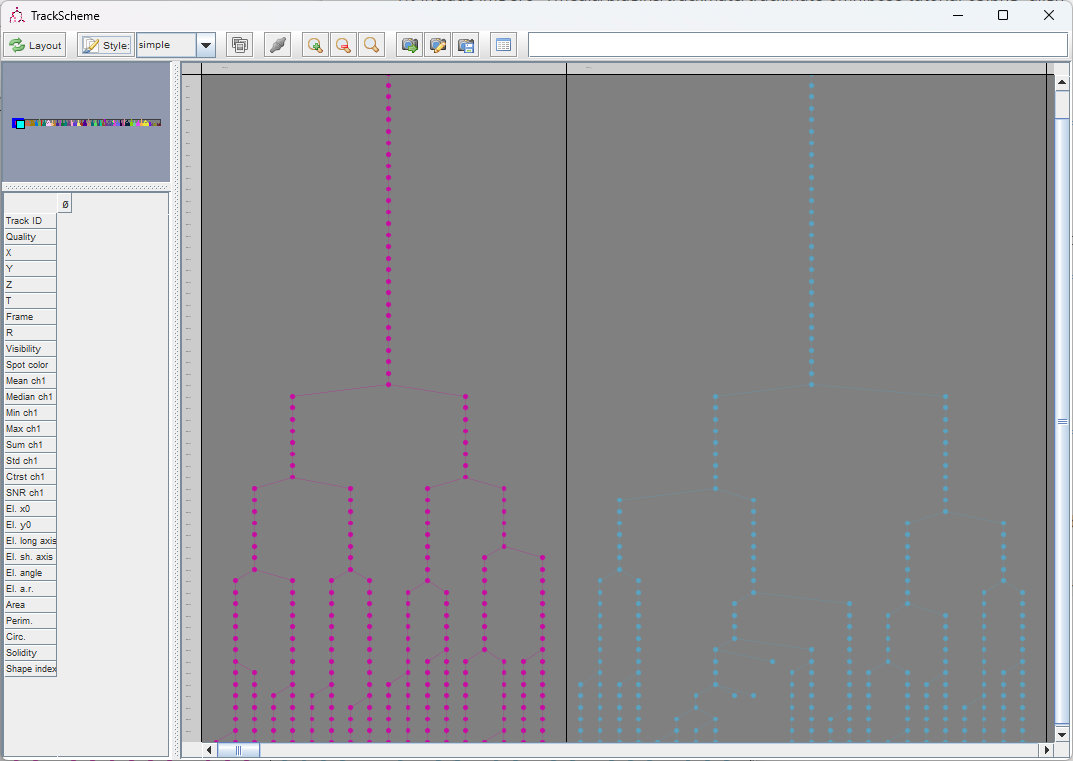
The lineages that we see in the TrackScheme window show that for some bacteria, there might be some linking errors late in the movie. But overall we could correctly detect the division events over 5 generations in this movie.
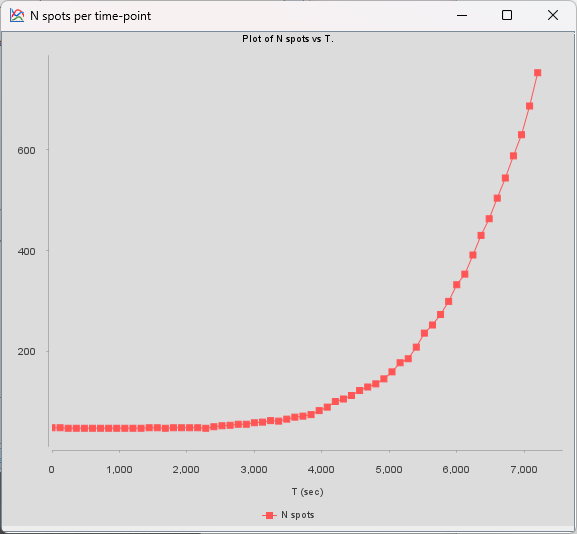
In the very last panel of TrackMate, an action called Plot N spots vs time highlights the exponential growth of the bacteria: