Please note that this version of the Stitching is deprecated. A new version is available here: Image Stitching
Citation
Please note that the Stitching, as well as other plugins available through Fiji, is based on a publication. If you use it successfully for your research please be so kind to cite our work:
- S. Preibisch, S. Saalfeld, P. Tomancak (2009) “Globally optimal stitching of tiled 3D microscopic image acquisitions”, Bioinformatics, 25(11):1463-1465. Webpage PDF
Introduction
There is an increasing demand to image large biological specimen at high resolution. Typically those specimen do not fit in the field of view of the microscope. To overcome this drawback, motorized stages moving the sample are used to create a tiled scan of the whole specimen. The physical coordinates provided by the microscope stage are not precise enough to allow reconstruction (“Stitching”) of the whole image from individual image stacks.
The 2D/3D Stitching Plugin is able to reconstruct big images/stacks from an arbitrary number of tiled input images/stacks, making use of the Fourier Shift Theorem that computes all possible translations (x, y[, z]) between two 2D/3D images at once, yielding the best overlap in terms of the cross correlation measure. If more than two input images/stacks are used the correct placement of all tiles is determined using a global optimization. To remove brightness differences at the tile borders, non-linear intensity blending is applied.
Overview of the Stitching Plugins
The Stitching package comes with 4 different plugins:
- 2D Stitching: Stitch two 2-dimensional images, rectangular ROIs can be used to limit the area to search in.
- 3D Stitching: Stitch two image stacks (3D), rectangular ROIs can be used to limit the area to search in as well.
- Stitch Grid of Images: Stitch an arbitrary number of 2D or 3D images which are arranged in a regular grid (e.g. 4 x 3 images with roughly 10% overlap)
- Stitch Image Grid Sequence: Stitch a sequence of 2D or 3D regular grids of images.
- Stitch Collection of Images: Stitch an arbitrary number of 2D or 3D images which do not have to be in a regular grid. The rough initial configuration (which image is overlapping another one) is given in tile configuration file.
- Stitch Directory with Images (unknown configuration)): Stitch all images that are located in a given directory without any knowledge of the tile configuration.
2D Stitching and 3D Stitching
The plugins 2D/3D Stitching take two 2D/3D images as input. They can contain rectangular ROIs which limit the search to those areas, however, the full images will be stitched together. If the input images are RGB images the user can select which channels will be used to determine the correct overlap. Note: If the stitching was not successful using ROIs on the obviously overlapping area might solve the problem.
Windowing should be always used, except the user knows well about properties of the Fourier Transform and its drawbacks.
The number of peaks defines the number of maxima in the Phase Correlation Matrix which are examined. If the stitching was not correct increasing this number might solve it.
There are 5 fusion methods available right now.
- Average: In the overlapping area the average intensity between both images is computed.
- Linear blending: In the overlapping area the intensity are smoothly adjusted between the two images. The fusion alpha defines the blending function ( alpha = 0 blending by averaging, alpha = 1 linear blending, alpha > 1 non-linear blending ).
- Max. Intensity: In the overlapping area the maximum intensity between both images is used int the output image.
- Min. Intensity: In the overlapping area the minimum intensity between both images is used int the output image.
- Red/Cyan Overlay: In the output image the first image is colored in red, the second one colored in cyan to visualize the registration. In the overlapping area the intensities should appear in gray scales (R + GB). This fusion method does not work with RGB input images for obvious reasons.
Stitch Grid of Images
This plugin is able to stitch a grid of images, it does not matter if it is 2D or 3D images as long as all images are of the same type. In contrast to the 2D/3D stitching of two images, this plugins will load the images from harddisc.
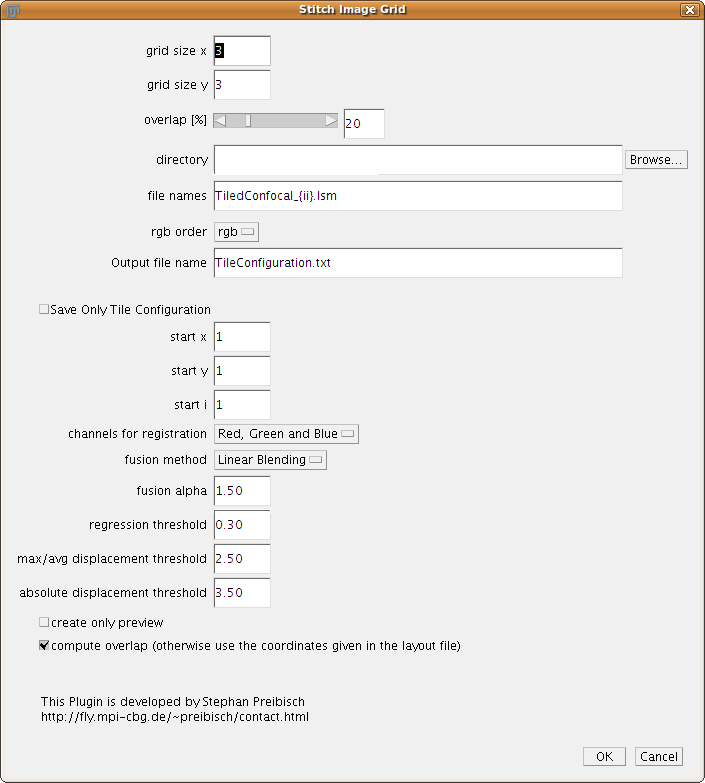
The user has to define the grid size, that means how the input tiles are arranged (e.g. 7 x 7 image tiles). The overlap is a rough estimate. Note: Smaller overlap reduces computation time, but if the correct alignment is not found try increasing this value first.
The next entries in the dialog are used to tell the plugin how to load the images which are in the grid. The directory should point to the directory where the images are stored. In the file names the user has to tell the program how the files are named (e.g. 49 different file names). There are typically two ways how the files are named:
- Images have an increasing number:
Tile_001.lsmTile_002.lsm….Tile_049.lsm - Images have the x and y coordinate in their filenames:
Tile_01_01.tifTile_01_02.tif….Tile_07_07.tif
These two cases would be described as follows:
- Images have an increasing number:
Tile_{iii}.lsm- The index number has always three digits and is just counted up starting at 1. The plugin knows that it has to count till 49 as there are 7 x 7 tiles. - Images have the x and y coordinate in their filenames:
Tile_{xx}_{yy}.tif- The index numbers for x and y coordinate of the tile have always two digits and are counted up starting at one each. The plugin knows that it has to count to 7 each as there are 7 x 7 tiles.
The starting numbers for x, y and i can be changed in the boxes below. The plugin will write a tileconfiguration.txt into the directory where the files are stored which can be investigated and changed if wanted. The plugin Stitch Collection of Images will take such a file as input. If you hit the checkbox “Save only tile configuration” the plugin will just create this file and stop.
The RGB order can be used to define how the different channels (if there are more than one) are read into the channel Red Green and Blue.
The channels for registration as well as the fusion methods work the same as described above in the 2D and 3D Stitching Plugin section. Note: Maximum intensity fusion is the most memory efficient stitching as it does not need to hold all images in memory at once!
The last three entries describe the behaviour of the global optimization:
- regression threshold: If the regression threshold between two images after the individual stitching are below that number they are assumed to be non-overlapping. Typical values for a good registration are over 0.7, however in noisy images with less content also small regression thresholds can be a correct overlap.
- max/avg displacement threshold: After the overlap between all individual tiles has been computed the global optimization computes an optimal arrangement of all tiles. After that, some image pairs will be placed different to each other compared to the individual alignment, which we call displacement. If there are no major alignment errors the average and maximal displacement will be below or around 1 pixel. If one of the individual alignment between two images was wrong, this pair will be displaced a lot in the global optimization as all other connecting tiles pull it towards the correct global position. If the maximal displacement is much higher than the average one it means that this individual alignment is most likely wrong and will be removed. Note: An image will just be removed if there is no link to another image any more.
- absolute displacement threshold: Removes links between images if the absolute displacement is higher than this value.
If you check the Create only Preview checkbox the program will not register the input data but only show a the principal layout of the files to check if your input was correct.
Stitch Image Grid Sequence
This plugin complements the previous one by stitching a sequence of grid of image that must have the same X-Y grid configuration. Apart from the previous parameters, the user must now select the number of sections (grid size z), the file name template (for example Tile_Z{zzz}_Y{yyy}_X{xxx}.lsm), the starting z value and the output directory where all resulting stitched images will be stored.
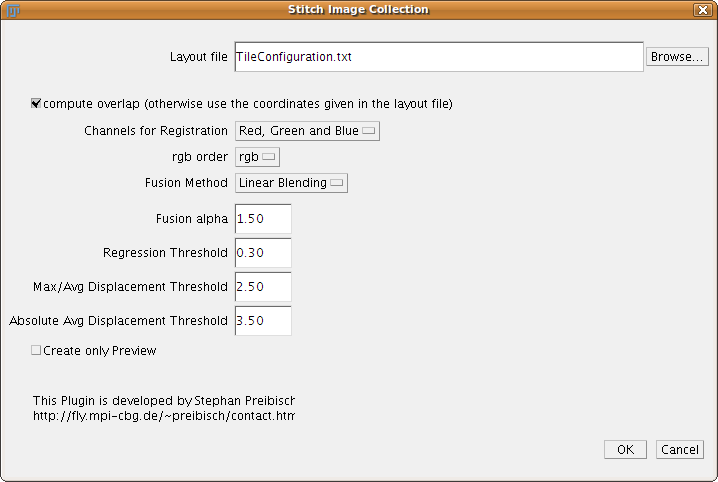
Stitch Collection of Images
This plugin takes an arbitrary tile configuration as input which has to be stored in a file.
The plugin Stitch Grid of Images will for example create such a file.
All the other parameters are the same as explained in the section Stitch Grid of Images.
Stitch Directory with Images (unknown configuration)
This plugin takes all images that are located in a given directory and tries to align them without any knowledge of the tile configuration. All other parameters are identical to those explained in the section Stitch Grid of Images. If the plugin does not find the correct alignment, you can try to increase the cross correlation threshold so that only pairwise alignments with very high quality are used for the global optimization. This increases the chance that the optimizer is able to detect the correct pairwise alignments. This, however, requires images with high quality as otherwise correct pairwise alignments with relatively low correlation are excluded.
Stitch Multiple Series or Tile Scan File
This plugin takes series of images contained in a single file, e.g. in the .lsm file format. It relies on Bio-Formats’s series support. The parameters are as explained in the section Stitch Grid of Images.
Problems and Known Issues
If the Stitch Grid, Stitch Collection or Stich with Unknown Configuration plugins are not able to create the correct output image you can do it yourself iteratively using the 2D/3D Stitching plugins. You just start with the first two images and fuse them. Using ROIs on clearly similar areas you can force a correct alignment. Afterwards you fuse the result with the third image and so on. The biggest drawback apart from the time consumption is that you can only use Maximum Intensity as fusion method, otherwise the image will look weird. Furthermore it will consume more memory.
For loading microscopic images we use the LOCI Bioformats importer. If you experience problems loading files, convert them to TIFF before, it should be read fine.
3D Stitching will not work for z-Stack size of smaller than 3 pixels. If you want to reconstruct such image from very thin tiles, duplicating of some of the stacks should solve the problem.
Results & Computation time
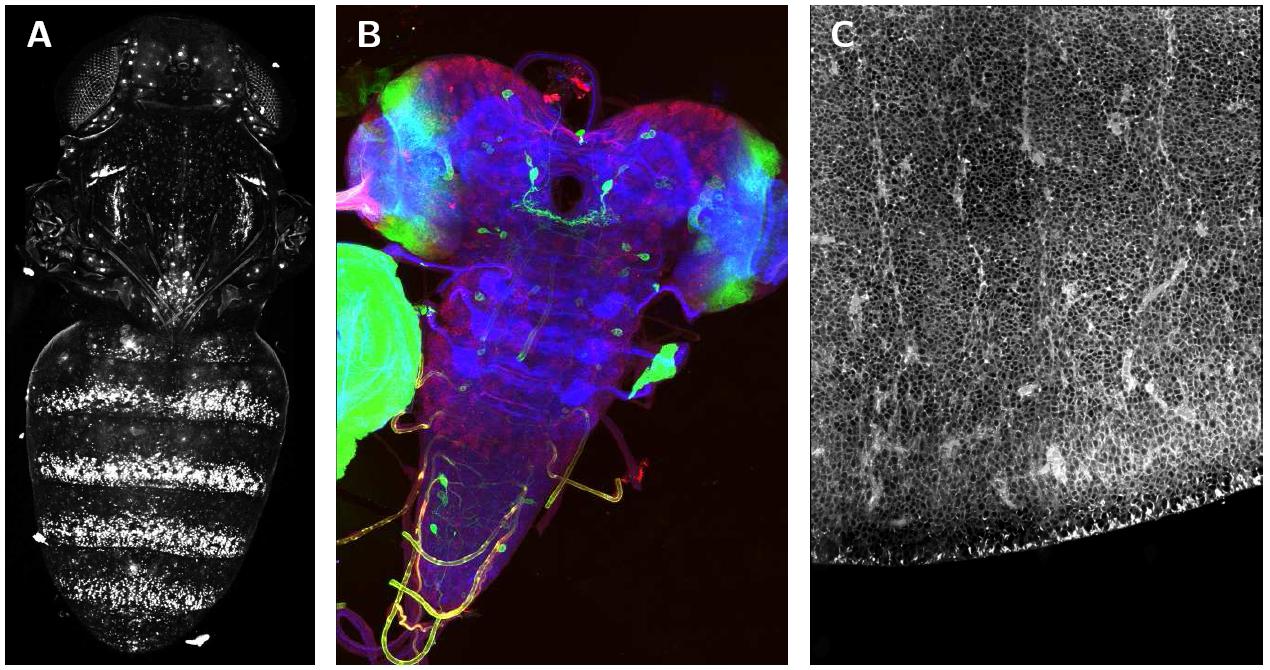
The figure shows stitched images of 3D confocal tiles. (A) shows a Drosophila melanogaster pupae expressing a GFP reporter under the regulation of the yellow gene, imaged few hours before eclosion using a 4× dry lens on an Optiphot confocal microscope (Nikon). It was stitched from three image stacks arranged in a 1 × 3 grid (Table 1 first row). The maximum intensity projection is shown. (B) shows the Drosophila larval nervous system stained with three dyes, stitched from a grid of 2 × 3 RGB images (see table 1 second row), the maximum intensity projection is shown. (C) shows a zone in the dorsal telencephalon of human embryonic tissue from week 17 post conception, incubated for 24 hours at 37°C in DiI. It was imaged using a 63×/1.4 objective on Zeiss LSM 510 equipped with a motorized stage. The final image was created from 24 image stacks arranged in a 4 × 6 grid (see table 1 third row), slice 18 is shown. Special thanks to Nicolas Gompel, James W. Truman, Simone Fietz and Wieland B. Huttner for providing the images.
For interactive examples of these datasets have a look here.
| tiles | individual tile dimension | output image dimension | output image size | computation time | min/avg/max displacement |
|---|---|---|---|---|---|
| 3 | 1024×1024×42 | 1097×2345×43 | 108 MB (8 Bit) | 0:42 min | 0.00/0.00/0.00 px |
| 6 | 512×512×86 | 975×1425×86 | 350 MB (RGB) | 1:20 min | 0.60/0.77/1.05 px |
| 24 | 1024×1024×68 | 3570×5211×70 | 1200 MB (8 Bit) | 22:43 min | 0.49/0.76/0.99 px |
| 72 | 512×512×122 | 3391×3847×145 | 1850 MB (8 Bit) | 43:10 min | 0.00/0.39/0.64 px |
| 63 | 1024×1024×92 | 6088×7667×119 | 5424 MB (8 Bit) | 178:57 min | 0.00/0.66/1.18 px |
The Table shows examples of stitched data computed on an Intel® Quad-Core CPU machine with 2.67GHz and 24GB of RAM. The global alignments of all stitchings have an average error below 1 px, the displacements in row 1 are zero because the two alignments are independent of each other. Note that the computation time scales roughly linearly with the output image size.
Contact
For any type of comment, questions or input please write to preibisch@mpi-cbg.de or visit my homepage.
See Also
- The Publication on the Stitching Plugin, S. Preibisch, S. Saalfeld, P. Tomancak (2009) Globally optimal stitching of tiled 3D microscopic image acquisitions”, Bioinformatics, 25(11):1463-1465. PDF
- TrakEM2 for non-destructive stitching with floating, adjustable images.
- XuvTools similar stitching software from the University of Freiburg and the abstract of the accompying publication