The content of this page has not been vetted since shifting away from MediaWiki. If you’d like to help, check out the how to help guide!
Do you need image data to try your algorithms on? Do you lack expert ground truth to test your methods? No problem! Here you have a list of available public data sets from the Fiji community and other sources:
We provide two image stacks where each contains 20 sections from serial section Transmission Electron Microscopy (ssTEM) of the Drosophila melanogaster third instar larva ventral nerve cord. Both stacks measure approx. 4.7 x 4.7 x 1 microns with a resolution of 4.6 x 4.6 nm/pixel and section thickness of 45-50 nm.
In addition to the raw image data, we provide for the first stack a dense labeling of neuron membranes (including orientation and junction), mitochondria, synapses and glia/extracellular space. The first stack serves as a training dataset, and a second stack of the same dimension can be used as a test dataset.
All the data and further information is available in the GitHub repository.
- Segmented ssTEM stack of neural tissue, thanks to Albert Cardona.
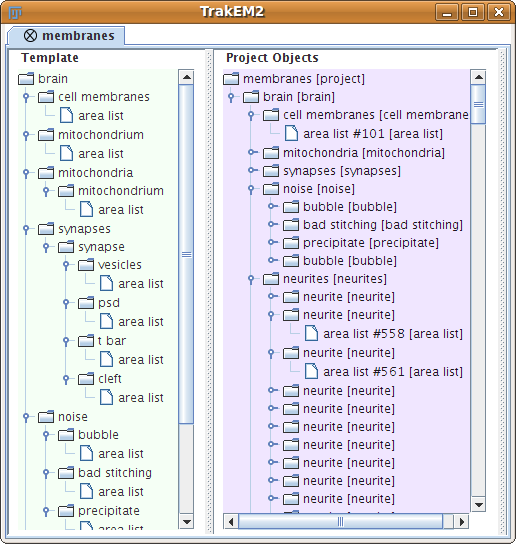

30 sections from a serial section Transmission Electron Microscopy (ssTEM) data set of the Drosophila first instar larva ventral nerve cord (VNC). The microcube measures 2 x 2 x 1.5 microns approx., with a resolution of 4x4x50 nm/pixel.
The challenge: use this data set to train machine learning software for the purpose of automatic segmentation of neural structures in ssTEM. More information in the 2012 open competition page.
The images are representative of actual images in the real-world: there is a bit of noise; there are image registration errors; there is even a small stitching error in one section. None of these led to any difficulties in the manual labeling of each element in the image stack by an expert human neuroanatomist. A software application that aims at removing or reducing human operation must be able to cope with all these issues.
See other manually segmented serial section TEM data sets.
- Sample data at LOCI, in a variety of file formats.
- Sample OME-TIFF data on openmicroscopy.org, thanks to Josh Bembenek.
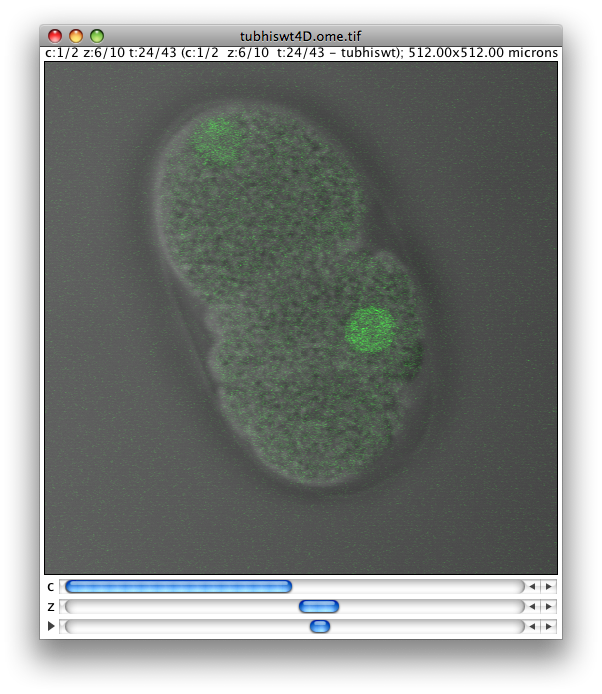
The dataset consists of tubulin histone GFP coexpressing C. elegans embryos. All image planes were collected at 512x512 resolution in 8-bit grayscale.
- Migrating macrophages in response to stimuli.
4D data set, kindly provided by Dirk Sieger and Francesca Peri, EMBL.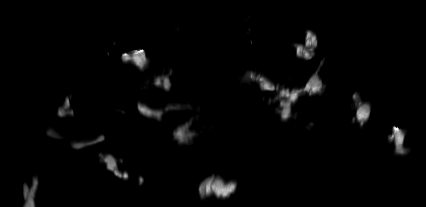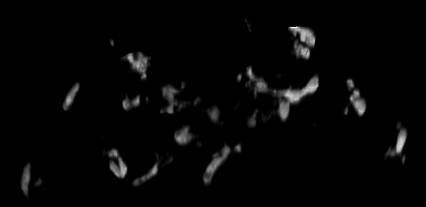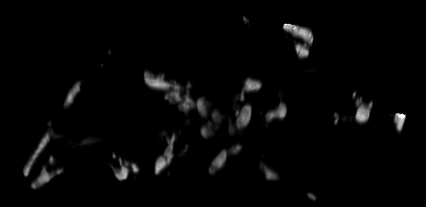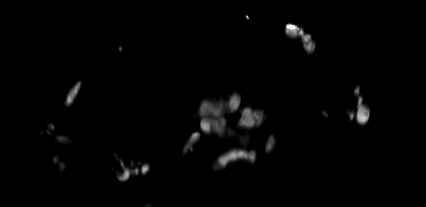
The challenge: trace the macrophages in 4D, and measure their shape volumes, surfaces, positions and pixel value intensities.
The file is a tif hyperstack that can be loaded directly into the 3D Viewer.
The key idea is to setup the tracking system so that it requires minimal user interaction, and is general. For example, a user clicks on one cell in one time point, and the program should find all other time points of the same cell. Then the program should learn about the statistics of that cell, and offer the means to automatically find all other cells in the volume and track them as well. In summary, a machine learning approach to 4D tracking.
- The Cell Image Library has many images related to cell biology; the images come with one of 4 licenses: Public Domain, Creative Commons Attribution, Creative Commons Attribution Non-Commercial Share-Alike, and Copyrighted (you need to ask the contributor whether you may use their image explicitly). This website allows you to contribute your own data sets, too!
- Open Connectome Project hosts a 10 terabyte data set of the mouse visual cortex (Bock et al., 2011) with CATMAID, and will offer means to run arbitrary programs on the data very soon.
- A 3D electron microscopy dataset of rodent brain, courtesy of Graham Knott and Marco Cantoni at EPFL. In particular, a FIBSEM volume measuring 5x5x5 micrometers and taken from the CA1 hippocampus region of the brain, and imaged at the extraordinary resolution of 5x5x5 nanometers per voxel! In addition, Aurelien Lucchi from Pascal Fua’s lab has made available, in the same page, image volumes containing labels for mitochondria.
- Broad Bioimage Benchmark Collection: annotated biological image sets for testing and validation. Collection of freely downloadable microscopy image sets. In addition to the images themselves, each set includes a description of the biological application and some type of “ground truth” (expected results).