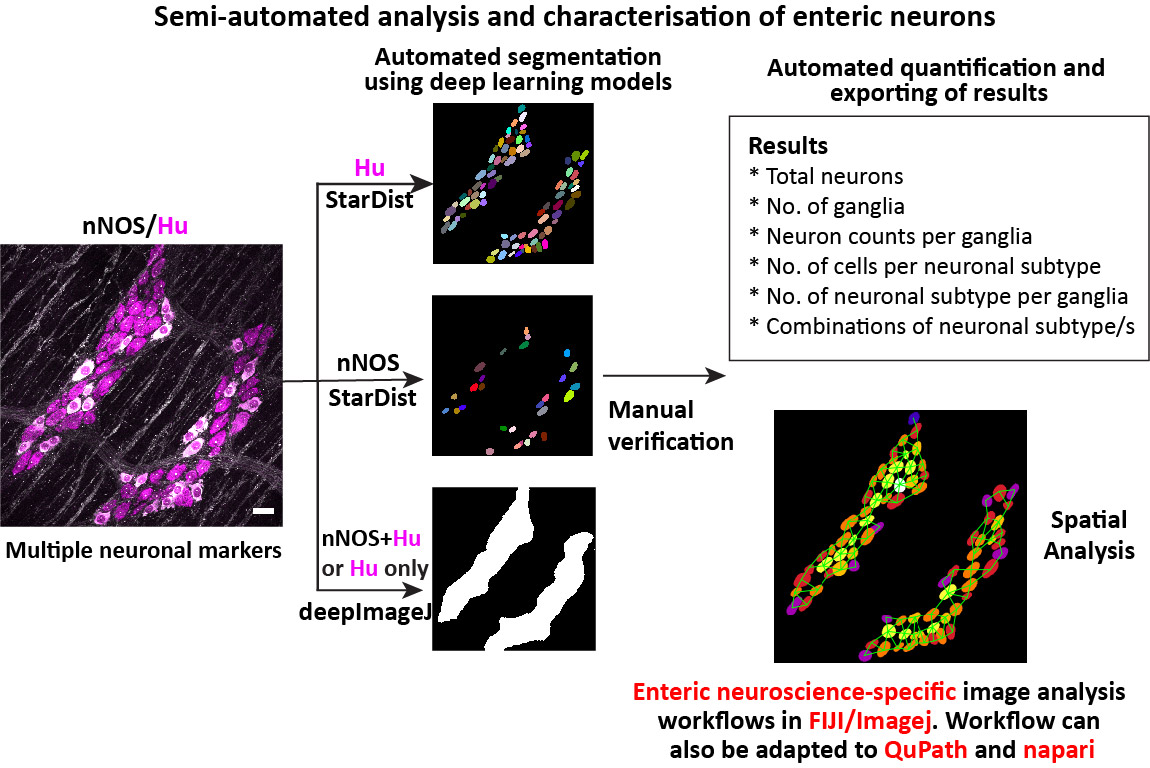
Gut Analysis Toolbox enables semi-automated analysis of the neuronal and glial distribution within the gut wall (enteric nervous system). It uses a combination of FIJI macros, StarDist, CLIJ
and deepImageJ for segmentation and analysis.
Click here to access the WIKI detailed instructions on how to use GAT.
You can also watch tutorials for GAT on Youtube.
What you can do with GAT:
- Semi-automated analysis of enteric neuronal distribution: Uses pan-neuronal marker Hu or any marker with similar labelling
- Normalise counts to the number of ganglia.
- Count number of neuronal subtypes, such as ChAT, nNOS etc..
- Spatial analysis with number of neighboring cells.
- Calcium imaging analysis: Alignment of images and extraction of normalised traces
- Multiplex image registration
Reference
To cite GAT:
Luke Sorensen, Adam Humenick, Sabrina S. B. Poon, Myat Noe Han, Narges S. Mahdavian, Matthew C. Rowe, Ryan Hamnett, Estibaliz Gómez-de-Mariscal, Peter H. Neckel, Ayame Saito, Keith Mutunduwe, Christie Glennan, Robert Haase, Rachel M. McQuade, Jaime P. P. Foong, Simon J. H. Brookes, Julia A. Kaltschmidt, Arrate Muñoz-Barrutia, Sebastian K. King, Nicholas A. Veldhuis, Simona E. Carbone, Daniel P. Poole, Pradeep Rajasekhar; Gut Analysis Toolbox: Automating quantitative analysis of enteric neurons. J Cell Sci 2024; jcs.261950. doi: https://doi.org/10.1242/jcs.261950
To download the training data, notebooks and associated models please go to the following Zenodo link: