Latest release: March 2019
About the plugin
SPIM Workflow Manager for HPC is a Fiji plugin developed at IT4Innovations, Ostrava, Czech Republic. The plugin enables biology users to upload data to the remote cluster, monitor computation progress, and examine and download results via the local Fiji installation.
Background
Imaging techniques have emerged as a crucial means of understanding the structure and function of living organisms in primary research, as well as medical diagnostics. In order to maximize information gain, achieving as high spatial and temporal resolution as practically possible is desired. However, long-term time-lapse recordings at the single-cell level produce vast amounts of multidimensional image data, which cannot be processed on a personal computer in a timely manner, therefore requiring utilization of high-performance computing (HPC) clusters. For example, processing a 2.2 TB dataset of drosophila embryonic development, taking a week on a single computer, was brought down to 13 hours by employing an HPC cluster supporting parallel execution of individual tasks [automated workflow]. Unfortunately, life scientists often lack access to such infrastructure.
Addressing this issue is particularly challenging as Fiji is an extraordinarily extensible platform and new plugins emerge incessantly. So far, plugin developers have typically implemented task parallelization within a particular plugin, but no universal approach has yet been incorporated into the SciJava architecture. Here we propose the concept of integrating parallelization support into one of the SciJava libraries, thereby enabling developers to access remote resources (e.g., remote HPC infrastructure) and delegate plugin-specific tasks to its compute nodes. As the cluster-specific details are hidden in respective interface implementations, the plugins can remain extensible and technology-agnostic. In addition, the proposed solution is highly scalable, meaning that any additional resources can be efficiently utilized.
Description
SPIM data processing pipeline
SPIM (“Selective/Single Plane Illumination Microscopy”) typically images living biological samples from multiple angles (views) collecting several 3D image stacks to cover the entire biological specimen. The 3D image stacks, representing one time point in a long-term time-lapse acquisition, need to be registered to each other which is typically achieved using fluorescent beads as fiduciary markers. After the registration, the individual views within one time point need to be combined into a single output image either by content-based fusion or multi-view deconvolution [Multiview-reconstruction]. The living specimen can move during acquisition, necessitating an intermediate step of time-lapse registration. Whereas parallel processing of individual time points has proven to be beneficial, the time-lapse registration takes only a few seconds and can therefore be performed on a single computing node without the need for parallelization.
The sheer amount of the SPIM data requires conversion from raw microscopy data to Hierarchical Data Format (HDF5) for efficient input/output access and visualization in Fiji’s BigDataViewer (BDV). BDV uses an XML file to store experiment metadata (i.e. number of angles, time points, channels etc.). Although the conversion to HDF5 is a parallelizable procedure, further updating the XML file downstream in the pipeline is not; and per-time point XML files have to be created and then merged after completion of the registration and fusion steps. Consequently, the parallel processing of individual time points on an HPC resource (conversion to HDF5, registration, fusion and deconvolution) is interrupted by non-parallelizable steps (time-lapse registration and XML merging).
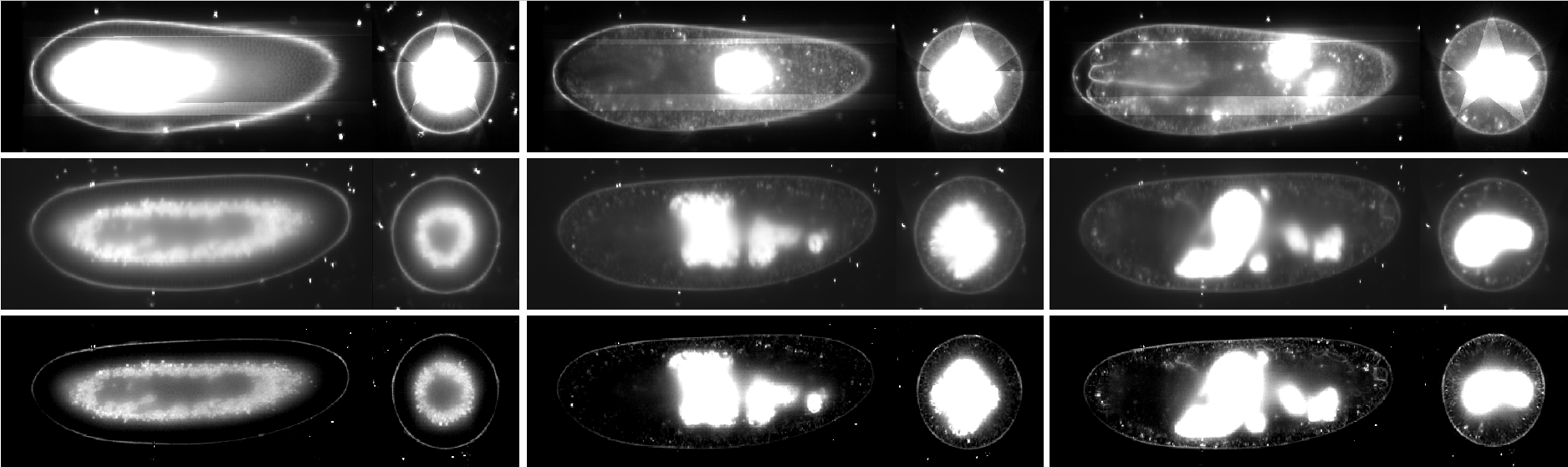
Pipeline input parameters are entered by a user into a config.yaml configuration file. In the first step, the .czi raw data are concurrently resaved into the HDF5 container in parallel on the cluster. Similarly, the individual time points are registered in parallel using fluorescent beads as fiduciary markers on the cluster. Subsequently, a non-parallel job executed by Snakemake consolidate the registration XML files into a single one, followed by time-lapse registration using the beads segmented during the spatial registration step. After this, the pipeline diverge into either parallel content-based fusion or parallel multi-view deconvolution. To achieve this divergence in practice, the Snakemake pipeline is launched from the Fiji plugin as two separate jobs using two different config.yaml files set to execute content-based fusion and deconvolution respectively. In the final stage of the pipeline, the fusion/deconvolution output is saved into a new HDF5 container. Figure below shows results of registration, fusion and deconvolution in different time points.
HEAppE middleware
Accessing a remote HPC cluster is often burdened by administrative overhead due to more or less complex security policies enforced by HPC centers. This barrier can be substantially lowered by employing a middleware tool based on the HPC-as-a-Service concept. To provide this simple and intuitive access to the supercomputing infrastructure an in-house application framework called High-End Application Execution (HEAppE) Middleware has been developed. This middleware provides HPC capabilities to the users and third-party applications without the need to manage the running jobs from the command-line interface of the HPC scheduler on the cluster.
To facilitate access to HPC from the Fiji environment, we utilize the in-house HEAppE Middleware framework allowing end users to access an HPC system through web services and remotely execute pre-defined tasks. Furthermore, HEAppE is designed to be universal and applicable to various HPC architectures. HEAppE also provides the mapping between the external users and internal cluster service accounts that are being used for the actual job submission to the cluster. It simplifies the access to the computation resources from the security and administrative point of view. For security purposes, users are permitted to run only a pre-prepared set of so-called command templates. Each command template defines an arbitrary script or an executable file which is to be run on the cluster, a set of input parameters modifiable at runtime, any dependencies or third-party software it might require, and the type of queue that should be used for the processing.
We developed a Fiji plugin underlain by HEAppE, which enables users to steer workflows running on a remote HPC resource. As a representative workflow we use a Snakemake based SPIM data processing pipeline operating on large image datasets. The Snakemake workflow engine resolves dependencies between subsequent steps and executes in parallel any tasks appearing to be independent, such as processing of individual time points of a time-lapse acquisition.
Installation
After you install and launch Fiji, go to Help › Update… › Manage update sites, tick HPC-ParallelTools and close the window. Then click Apply changes and restart Fiji.
Usage
Now you should see the plugin under Plugins › Multiview Reconstruction › SPIM Workflow Manager for HPC. Upon plugin invocation from the application menu, you are prompted for HEAppE credentials, e-mail address and specifying your working directory. Following a successful login, the main window containing all jobs arranged in a table is displayed. In this context, the term job is used for a single pipeline run with specified parameters. The plugin actively inquires information on the created jobs from HEAppE and updates the table as appropriate. 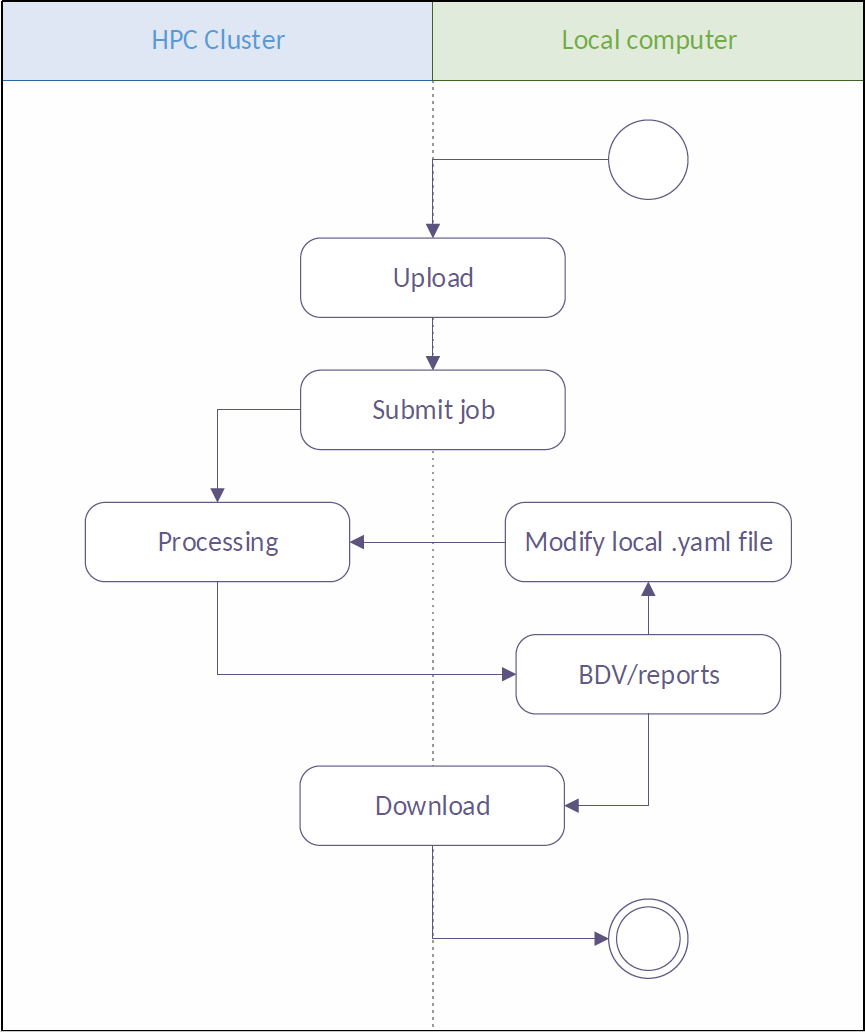
For creating a new job, right click in the main window and choose Create a new job. A window with input and output data location will pop up. You have the option to use demonstration data on the Salamon cluster or specify your own input data location. Eventually you may choose your working directory (specified during login) as both your input and output data location. Once a new job is configured, you are able to upload your own data (if you chose this option in the previous step) by right clicking on the job line and choosing Upload data. When Done appears in the Upload column, you can start the job by right click › Start job. Status of your job changes to Queued, then to Running and finally to Finished when your pipeline finishes successfully.
The plugin provides a wizard allowing you to set up a configuration file config.yaml, which effectively characterizes the dataset and defines settings for individual workflow tasks. The plugin supports uploading local input image data to the remote HPC resource, providing information on the progress and estimated remaining time.
Once a job execution is selected by you, the configuration file is sent to the cluster via HEAppE, which is responsible for the job life cycle from this point on. You can display a detailed progress dashboard showing current states of all individual computational tasks for the selected job as well as output logs useful for debugging.
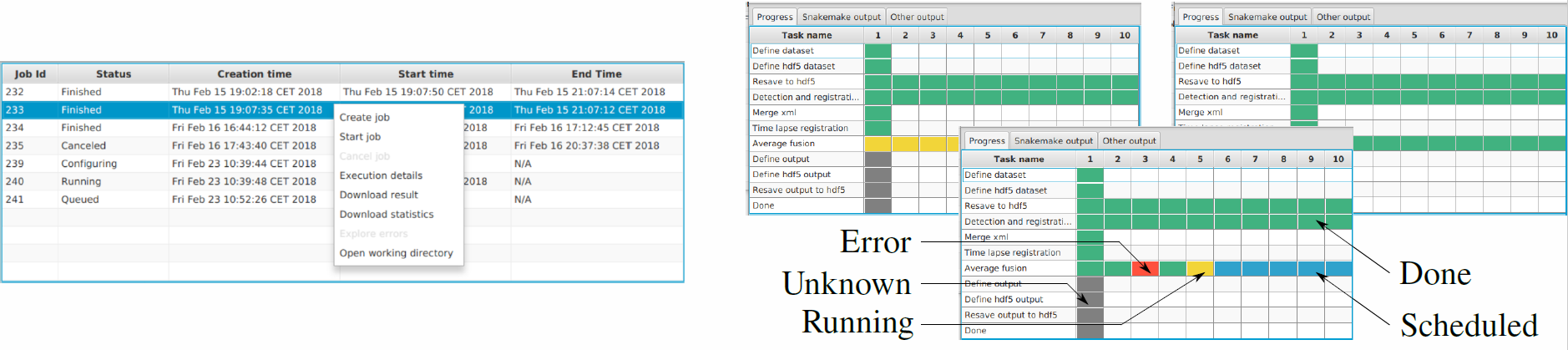
Following a successfully finished pipeline, you can interactively examine the processed SPIM image data using the BigDataServer as well as download resultant data and a summary file containing key information about the performed job. Importantly, you can edit the corresponding local configuration file in a common text editor, and restart an interrupted, finished, or failed job.
HPC Cluster
Execution of the Snakemake pipeline from the implemented Fiji plugin was tested on the Salomon supercomputer, at IT4Innovations in Ostrava, Czech Republic, which consists of 1 008 compute nodes, each of which is equipped with 2x12-core Intel Haswell processors and 128 GB RAM, providing a total of 24 192 compute cores of x86-64 architecture and 129 TB RAM. Furthermore, 432 nodes are accelerated by two Intel Xeon Phi 7110P accelerators with 16 GB RAM each, providing additional 52 704 cores and 15 TB RAM. The total theoretical peak performance reaches 2 000 TFLOPS. The system runs a Red Hat Linux.
Using the developed plugin, we executed the pipeline on the Salomon supercomputer. As the test data set we used 90 time-point SPIM acquisition of a Drosophila melanogaster embryo expressing FlyFos fluorescent GFP fusion reporter for the nrv2 gene. The embryo was imaged with Lightsheet Z.1 SPIM microscope (Carl Zeiss Microscopy) from 5 views every 15 minutes from cellular blastoderm stage until late stages of fruitfly embryogenesis. The dataset consisted of 170 GB of image data.
The data transfer and pipeline execution on Salomon using 90 nodes took 9 hours 37 minutes. For comparison, processing of the same dataset on a common workstation took 23 hours 56 minutes. The results show that despite the data transfer overhead, a significant speedup of SPIM image analysis has been achieved by employing HPC resources.
Citation
Please note that the plugin SPIM Workflow Manager for HPC available through Fiji is based on a publication. If you use it successfully for your research please be so kind to cite our work:
Jan Kožusznik, Petr Bainar, Jana Klímová, Michal Krumnikl, Pavel Moravec, Václav Svatoň, Pavel Tomančák; SPIM Workflow Manager for HPC, Bioinformatics, btz140, https://doi.org/10.1093/bioinformatics/btz140