The content of this page has not been vetted since shifting away from MediaWiki. If you’d like to help, check out the how to help guide!
Purpose
The ParticleSizer script was developed to automatically measures the distributions of the characteristic size and shape properties of a nanomaterial. In the scope of implementing the European Commission definition of a nanomaterial, the minimal external dimension of the primary particles of a particulate material is assessed as the minimal feret diameter from electron microscopy images. Other size and shape parameters are measured simultaneously.
Installation
The ParticleSizer script combines a number of different plugins. The most easy way (and at the moment the only way) is to follow Biomedgroup & ParticleSizer update site.
It is recommended to install R to get better plots. When this point is skipped, a stripped-down plot will be shown:
You can now start the ParticleSizer via Plugins › NanoDefine - › ParticleSizer
- Download the latest R 3.6 release: https://cran.r-project.org/bin/windows/base/old/3.6.3/
- Download the Rserver package: https://ftp.gwdg.de/pub/misc/cran/bin/windows/contrib/3.6/Rserve_1.7-3.1.zip
- Download the MASS package: https://ftp.gwdg.de/pub/misc/cran/bin/windows/contrib/3.6/MASS_7.3-54.zip
- Start R and select the packages downloaded in 1. and 2. via Start Packages › Install packages from local zip files
- Open Fiji, start the ParticleSizer Settings Manager and set the field
Set R install directoryto the R installation directory.
You can now start the ParticleSizer via Plugins › NanoDefine - › ParticleSizer
Settings
Segmentation: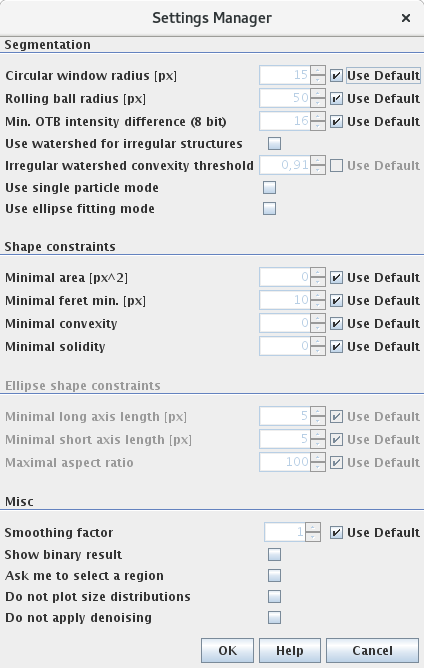
- Circular window radius: This is a parameter of the local thresholding technique. The ParticleSizer does not use a global threshold to binarize the image. Instead it uses a local threshold which is estimated for a specific circular region with the configured radius.
- Rolling ball radius: The background is removed by rolling a ball with this radius over the surface (intensity interpreded as hight) of the image. It should be at least as large as the largest object in image which does not belongs to the background.
- Min. OTB intensity difference: Objects which have an object-to-background (OTB) intensity difference in the noise-reduced and background subtracted image lower than this threshold are considered as artefacts and are removed.
- Use watershed for irregular structures: If selected, the mode for irregular structures is used.
- Irregular watershed convexity threshold: The threshold determines, when splitted agglomerated is counted as “primary particle”. If the convexity of particle is greater than this threshold, the splitting is stopped for this object. If the convexity is smaller than this value, then the ParticleSizer tries to split the particle again
- Use single particle mode: If selected, the single particle mode is used.
- Use ellipse fitting mode: If selected, the ellipse fitting mode is used.
Shape constraints:
- Minimal area: Minimum area in pixels. Particles smaller than this threshold are removed.
- Minimal feret min: Minimal feret diameter in pixel. Particles smaller than this threshold are removed.
- Minimal convexity: The convexity is defined as the ratio of the perimeter of the convex hull of the particle and the perimeter of the particle. It lies between 0 and 1. The convexity increases with larger values. Particles smaller than this threshold are removed.
- Minimal solidity: Defined as the ratio of the particle area and the area of the convex hull of the particle. It lies between 0 and 1. The solidity increases with larger values. Particles smaller than this threshold are removed.
Ellipse shape constraints:
- Minimal long axis length: The length in pixels of the major direction of the fitted ellipse. Ellipses smaller than this threshold are removed.
- Minimal short axis length: The length in pixels of the minor direction of the fitted ellipse. Ellipses smaller than this threshold are removed.
- Maximal aspect ratio: Ratio of the length of major and minor axis. Ellipses with an aspect ratio larger than this value are removed.
Misc:
- Smoothing factor: It sometimes occurs that the estimated standard deviation of the noise is lower than the true value. The smoothing factor is a multiplicative factor for the estimated standard deviation.
- Show binary result: If selected the ParticleSizer shows the binary result.
- Ask me to select a region: If selected, the software allows you to select a specific region to analyse.
- Record process: If this checkbox is selected, a new image stack opens at the end of the analysis which contain images of every single step the particle
sizer made. If an image stack is analysed, than only the only the processing of the first image is recorded.
How to find optimal settings
We distinguish two particle types:
- E: Ellipsoidal particles
- I: Irregular but convex particles
These particles could show some overlapping. We destinguish four different classes:
- N: No overlapping
- T: Touching
- S: Slightly overlapping
- H: High or complete overlapping
Combining both information, we could give the following advice (as a rule of thumb):
- N+E or T+E -> Keep the default settings
- N+I or T+I or S+I -> Activate the irregular watershed
- S+E -> Activate ellipse fitting
- H -> Activate single particle mode
Use the ParticleSizer with the NanoDefiner e-tool
The ParticleSizer supports the NanoDefiner e-tool. After the analysis of a dataset is finished, you can select the results table and export the cumulative mass function of the feret min diamater by Results › Export CMF. The exported file can be uploaded to the NanoDefiner e-tool.
Examples
Gold 8 nm, Settings: Default
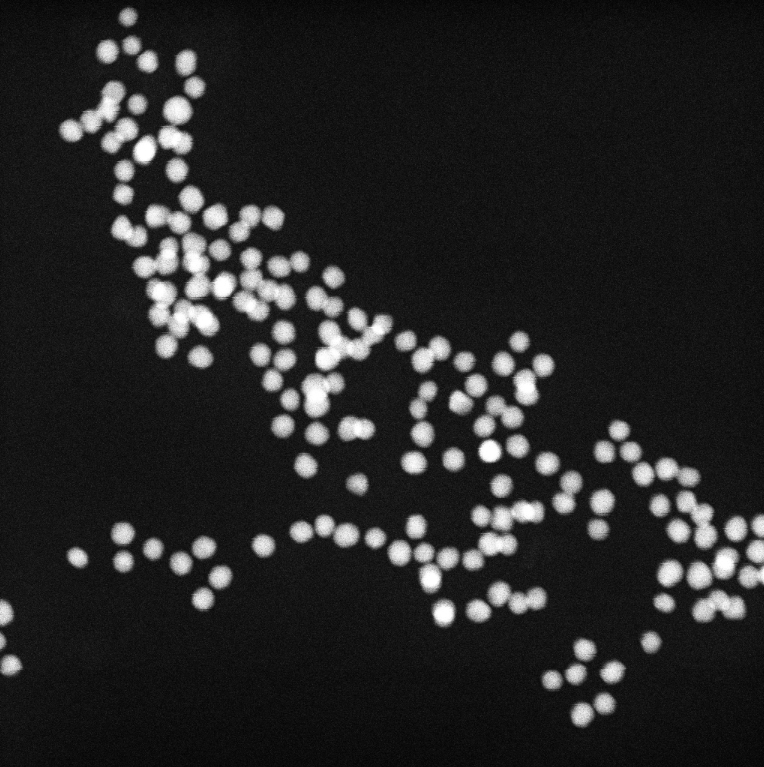
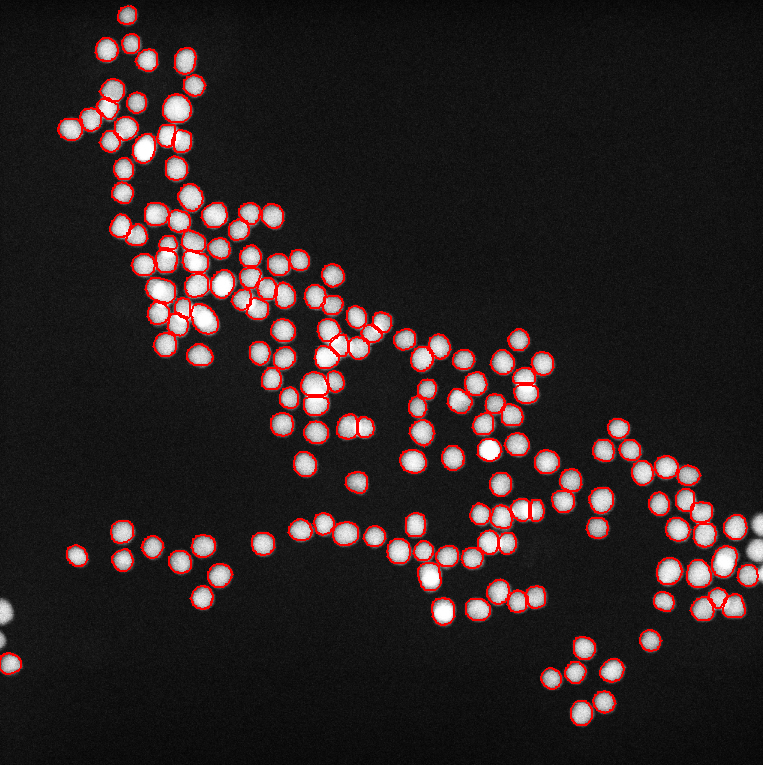
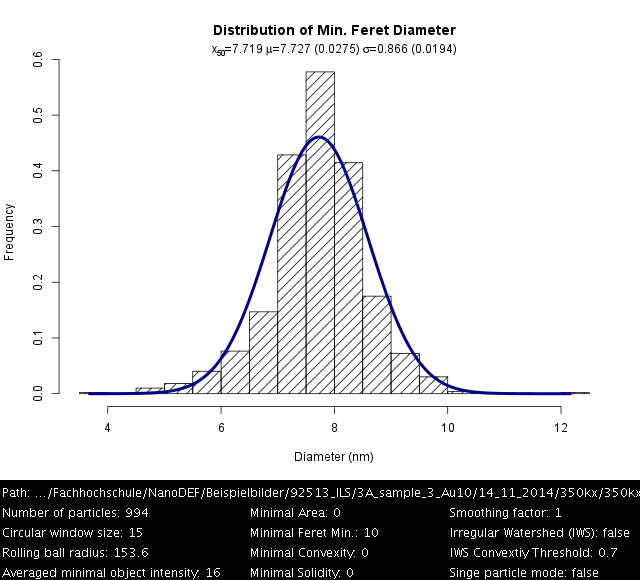
Gold 8 nm, Settings: +Ellipse fitting mode

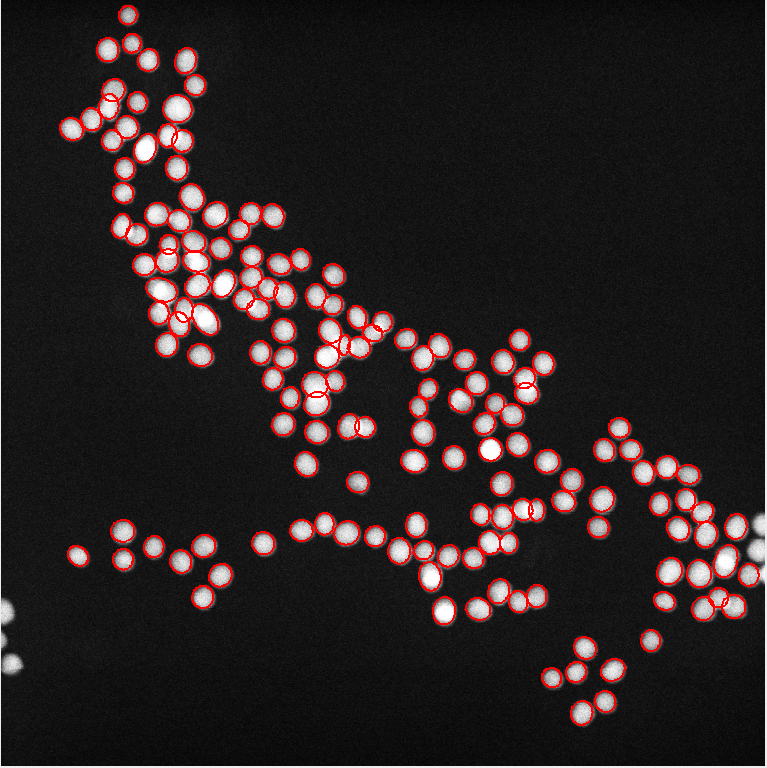
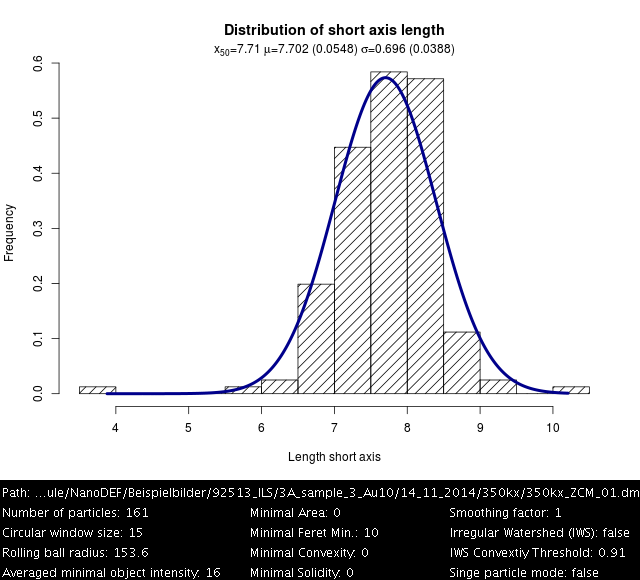
Gold 10 nm, Settings: +Min. OTB difference=30
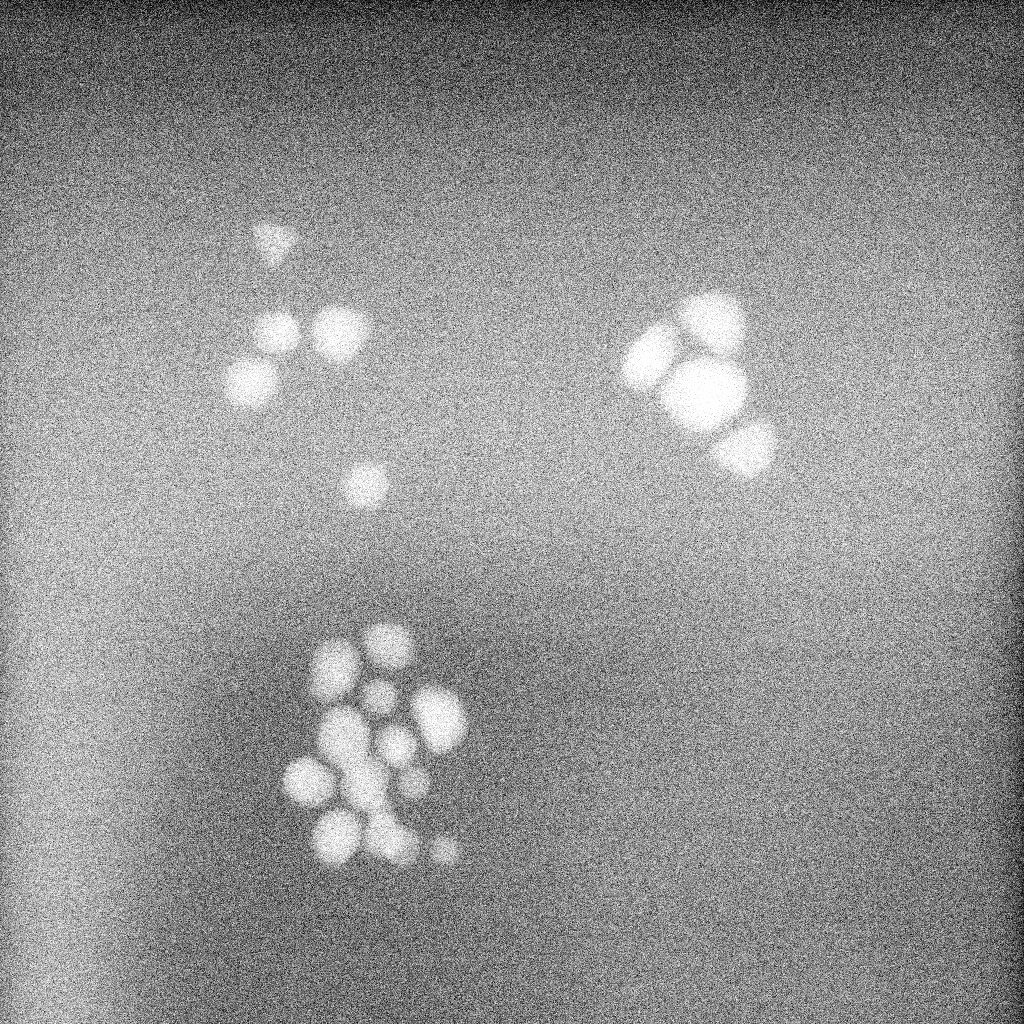

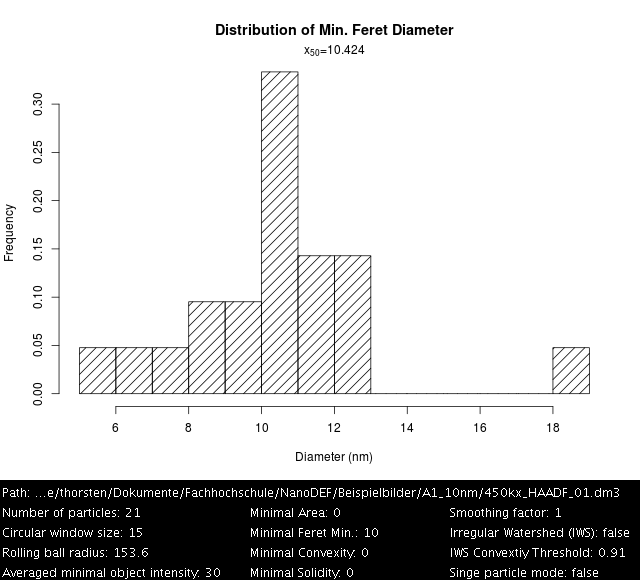
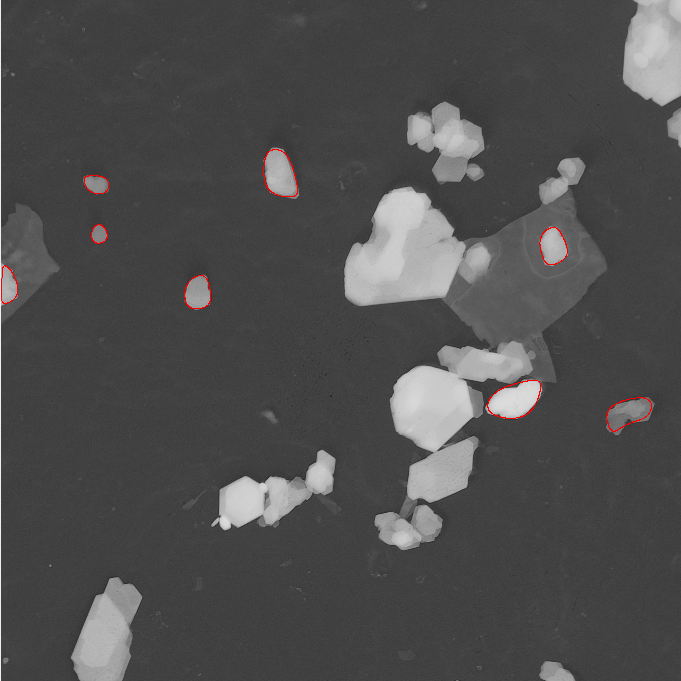
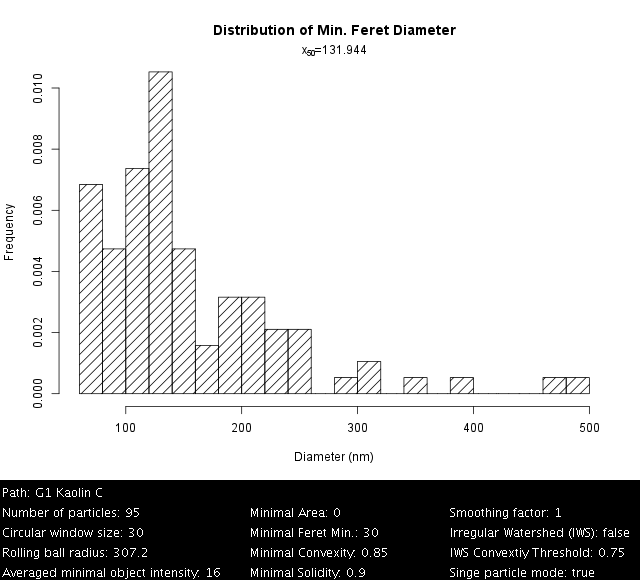
Kaolin, Settings: +Single particle mode

How to cite
A publication is in preparation.
You can use Zenodo to cite the lastest release of the ParticleSizer: https://zenodo.org/badge/latestdoi/18649/thorstenwagner/ij-particlesizer
As example: Thorsten Wagner. (2016). ij-particlesizer: ParticleSizer 1.0.1. Zenodo. 10.5281/zenodo.56457
Acknowledgements
The development of the ParticleSizer has received funding from the European Union Seventh Framework Programme (FP7/2007-2013)