The content of this page has not been vetted since shifting away from MediaWiki. If you’d like to help, check out the how to help guide!
|
|
|
| Category |
Segmentation |
Adiposoft is an automated Open Source software for the analysis of adipose tissue cellularity in histological sections. Adiposoft has been developed as a plug-in for Fiji (advanced distribution of ImageJ) that can be run under Windows, Linux or macOS. The software, that can be downloaded and used with no license restrictions, was developed at the Imaging Unit of the Center for Applied Medical Research (CIMA), University of Navarra.
A paper validating Adiposoft was published by Journal of Lipid Research in 2012.
Motivation
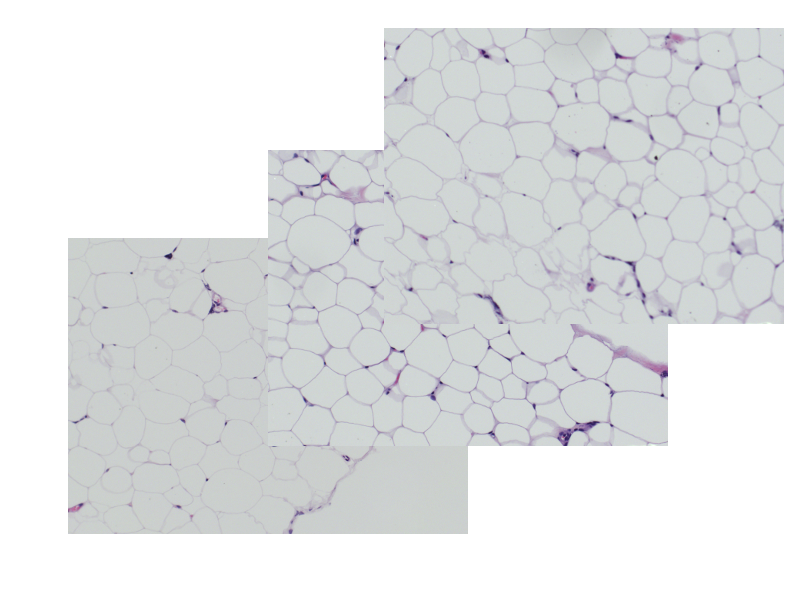
The accurate estimation of the number and size of adipocytes provides relevant information about the growth kinetics and the physiological status of a given tissue or organ. Adiposoft is an automated, open source software for the analysis of white adipose tissue cellularity on histological, hematoxylin and eosin (H&E) stained sections.
Download and Installation
The latest version of Adiposoft is 1.16. (Updated April 8th, 2019).
- If you do not have Fiji installed in your computer, you can download it here.
- Download the Adiposoft plugin here. Copy the Adiposoft jar file into the Plugins folder of Fiji application’s main folder. The Adiposoft tool will appear in the plugins menu the next time you start Fiji.
Running Adiposoft
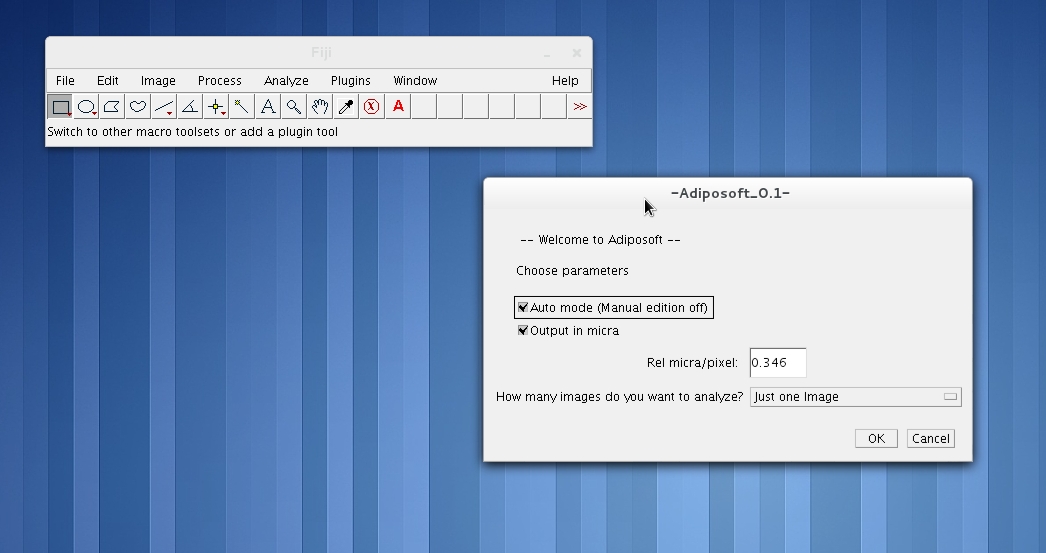
Go to the Plugins menu in Fiji, and click on Adiposoft. This will open the main Adiposoft dialogue window.
Selection of Parameters
On Adiposoft’s main dialogue window you can choose several parameters for the analysis. Namely:
- Auto mode: By checking on the “Auto mode” box, you choose to run Adiposoft in completely automated mode, without any user intervention, as it is described in the “Automated analysis” section below.
- Manual mode: If you uncheck the “Auto mode” box, you choose to have the opportunity to manually edit the results of the initial automated segmentation. This includes manually adding, deleting, splitting or merging adipocytes. The editor is activated after the initial automated analysis is done. This option is only available when the images are analyzed one by one (“One Image), and not in batch mode (“A Directory” or “A list of nested Directories – Batch Mode” ). The editing actions will be explained in the section “Manual analysis”.
- Output units. By default, the areas and diameters of the adipocytes are calculated and reported in pixels. If the calibration of the images is known, you can select “Microns”, but be ready to introduce the number of microns that correspond to one pixel of your images in the dialogue window that will appear when you press OK.
- How many images you want to analyze. You can choose to run Adiposoft to analyze one image at a time (“One Image”), a directory that contains all the images that you want to analyze (“A Directory”) or a directory and all directories below (“A list of nested Directories – Batch Mode”).
After pressing OK, you will be prompted to select the image that you want to analyze, the directory that contains the images -if you choose to analyze a “A Directory”- or the parent directory -if you choose to analyze “A list of nested Directories”-.
Next you will be prompted to choose –or create- the output directory where you want to store the results of the analysis.
Automated analysis
If you choose to run Adiposoft in Automated mode, the program analyzes the selected images, and stores the results in the output directory.
If you choose to analyze “One Image”, the output directory will contain an image with the results of the segmentation overlaid on the original image and an excel file containing the complete list of adipocytes with their area and equivalent diameter. If you choose to analyze “A Directory”, the output directory will contain an excel file with the list of adipocytes of all the images, with their area and parameters, and a directory named as the original input directory containing all the segmented images and one excel file per image, containing the results of that particular image. If you choose to analyze “A list of Directories”, the output folder will contain a tree of directories replicating the input tree, but containing the results of the analysis.
Manual analysis
If you choose to run Adiposoft in Manual mode, the program analyzes the selected image automatically and then offers the option to edit the results, using one of the editing options from a toolbar that appears in a new window. This toolbar contains the following buttons:
- End: Closes the window and stores the results in the selected output directory.
- Delete: Deletes an object, possibly corresponding to an incorrectly segmented adipocyte, or a tissue part incorrectly classified as being an adipocyte. When prompted, click on the object that you want to delete and press OK.
- Add: Allows drawing a closed contour that corresponds to the boundary of a cell that was not initially detected.
- Merge: Merges various objects, possibly parts of an incorrectly divided adipocyte. To this end, you can draw a closed contour. The final –merged- cell will contain –as a single object- the new area defined by the closed contour plus the areas of all cells touched by the contour.
- Separate: Divides an object in two parts, based on a line drawn by the user.
- Undo: Reverts the last action done.
Image DataSet
A set of images of histological sections obtained from three different rat tissue depots –retroperitoneal (RP), mesenteric (MES) and subcutaneous (SC)- can be downloaded and used to test Adiposoft. (Download)
Tools
Metamorph Macro (Molecular Devices, USA) that automated the acquisition of the images of the histological samples with the microscope.
Adiposoft under MATLAB (advanced users) - Maintained only until December 2014
A version of Adiposoft was entirely developed using MATLAB v.7.11 and the DIPlib v2.2 C Image Processing libraries [11] under Linux OS Fedora 14. To run Adiposoft, the same or newer version of this software is required, but the OS can be different. MATLAB requires a non-free license. ADIPOSOFT - MATLAB Version (Download)
Before installing Adiposoft under MATLAB you must download and install the DIP Image libraries DIPlib v2.2 C, which can be downloaded here. After installing DIPlib you must to extract the AdiposoftMatlab.zip file. To run Adiposoft, execute the .m file inside MATLAB
Note: Installing Adiposoft can be a bit tricky under MATLAB, due to compatibility problems between the versions of DIPlib and MATLAB. We will be pleased to help you with the installation. To that end, please contact mgalarraga@unav.es for help.