VTEA
| VTEA (Fiji) | |
|---|---|
| Author | Seth Winfree ([1]) |
| Maintainer | Seth Winfree ([2]) |
| Source | on GitHub |
| Development status | v0.7, alpha, 1.0.a on deck. |
| Category | Segmentation,Visualization,Plugins |
Contents
Volumetric Tissue Exploration and Analysis
Three-dimensional tissue cytometry. Three-dimensional tissue cytometry(3DTC) enables the quantitative measurement of whole cells while retaining their morphology, localization and associations-think of it as in situ flow cytometry. How can we do 3DTC? Volumetric tissue exploration and analysis!
In order for it to be useful it needs to be:
Free and easy to get. We opted to use ImageJ/FIJI as the distribution platform because it has an excellent and robust community of contributors. Practically, it provides the mechanisms for updating, a number of image processing tools and is built on a simple and powerful extensible framework.
Easy to use. We designed VTEA to organize the most common workflow in 3DTC inclusive of image processing (to manage imaging artifacts), segmentation (extensible to bring in edge deep learning approaches into a common framework of analysis) and exploration and analysis with flow cytometry like plots, gating, mapping to image with ROI gating and tools for high dimensionality data.
Mesoscale 3D analysis. We built segmentation to handle massive datasets and to operate seamlessly on an embedded database to enable analysis of 100’s of thousands of cells and will incorporate tools for clustering and dimensionality reduction.
Original image referencing. The power of 3DTC in VTEA enables the localization of identified cells in the analysis space in the original image, in situ, and in 3D with ClearVolume.
This brief video describes VTEA's core behaviors. VTEA's utility has been demonstrated in this paper.
We developed VTEA out of a need to unify the various tasks involved in image processing, segmenting, exploring and analyzing large 3D fluorescence light microscopy image volumes ranging from 50-100s of microns thick. Our solution is predicated upon the idea that image processing, segmentation and analysis of 3D image volumes is best implemented with a bidirectional interactive user interface from image processing to analysis.
Importantly, our solution is in its infancy. In fact, the tools are relatively simple ones drawn from ImageJ's core functions. These first tools represent only the beginning of our vision for VTEA. We are currently planning to leverage the SciJava framework to make VTEA easily extensible and continue to build both upon existing tools in ImageJ/Fiji and our own novel approaches.
Installing
To install the VTEA plugin use, How to follow a 3rd party update site and check the update site: "Volumetric Tissue Exploration and Analysis" OR follow the following four steps:
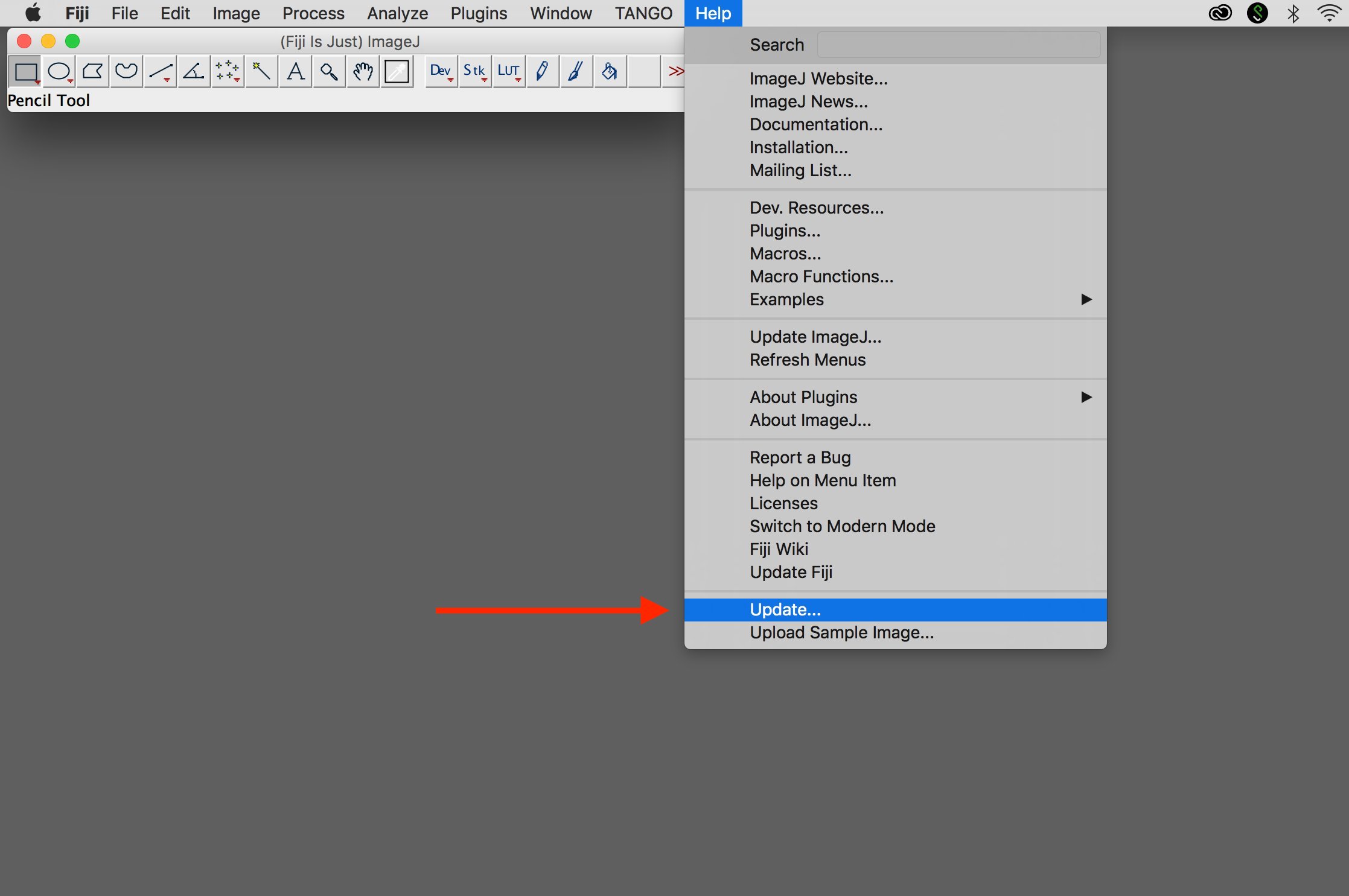
1. Start updater
Select "Update..." under "Help".
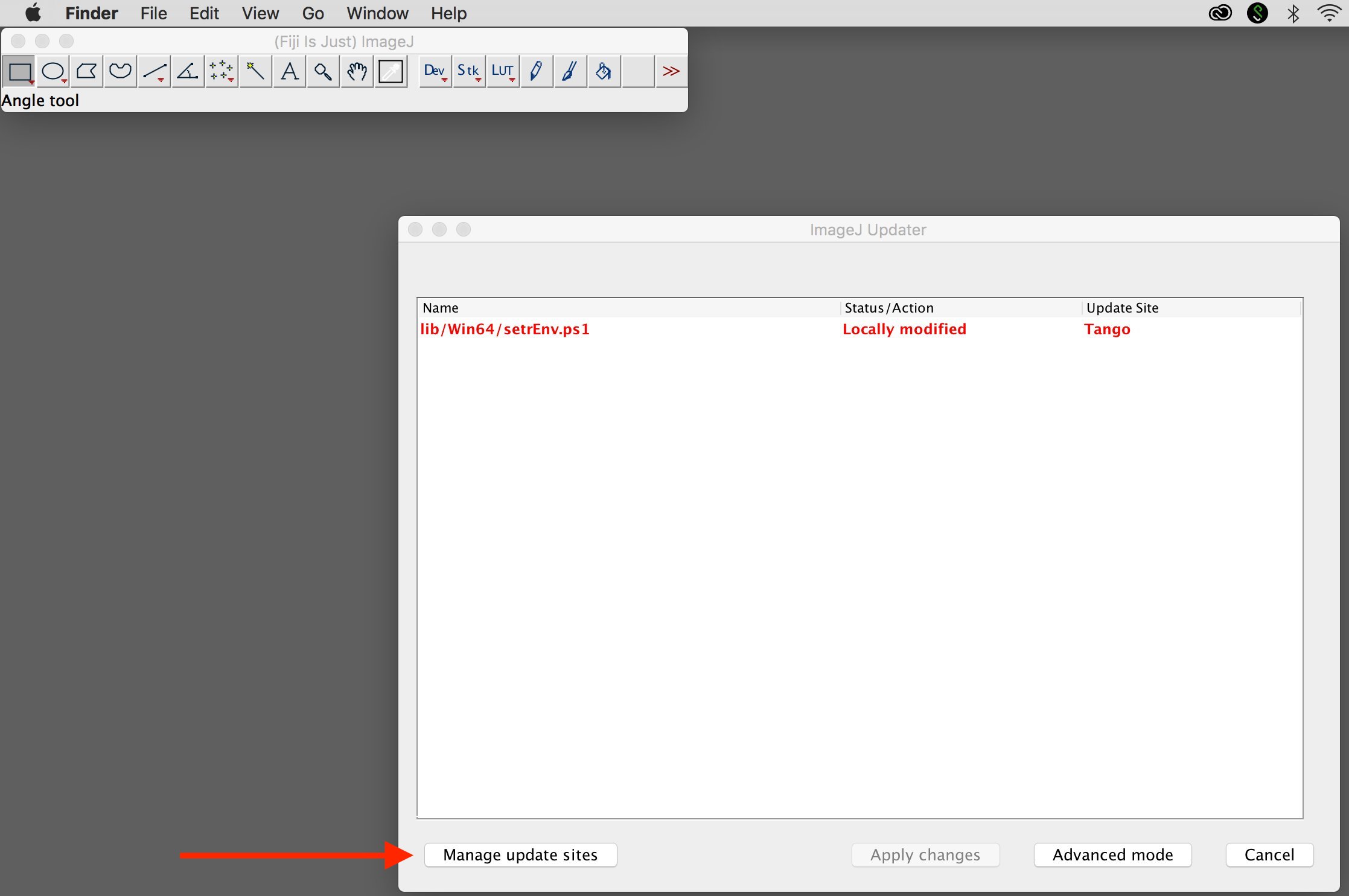
2. Add update site
Select "Manage update sites."
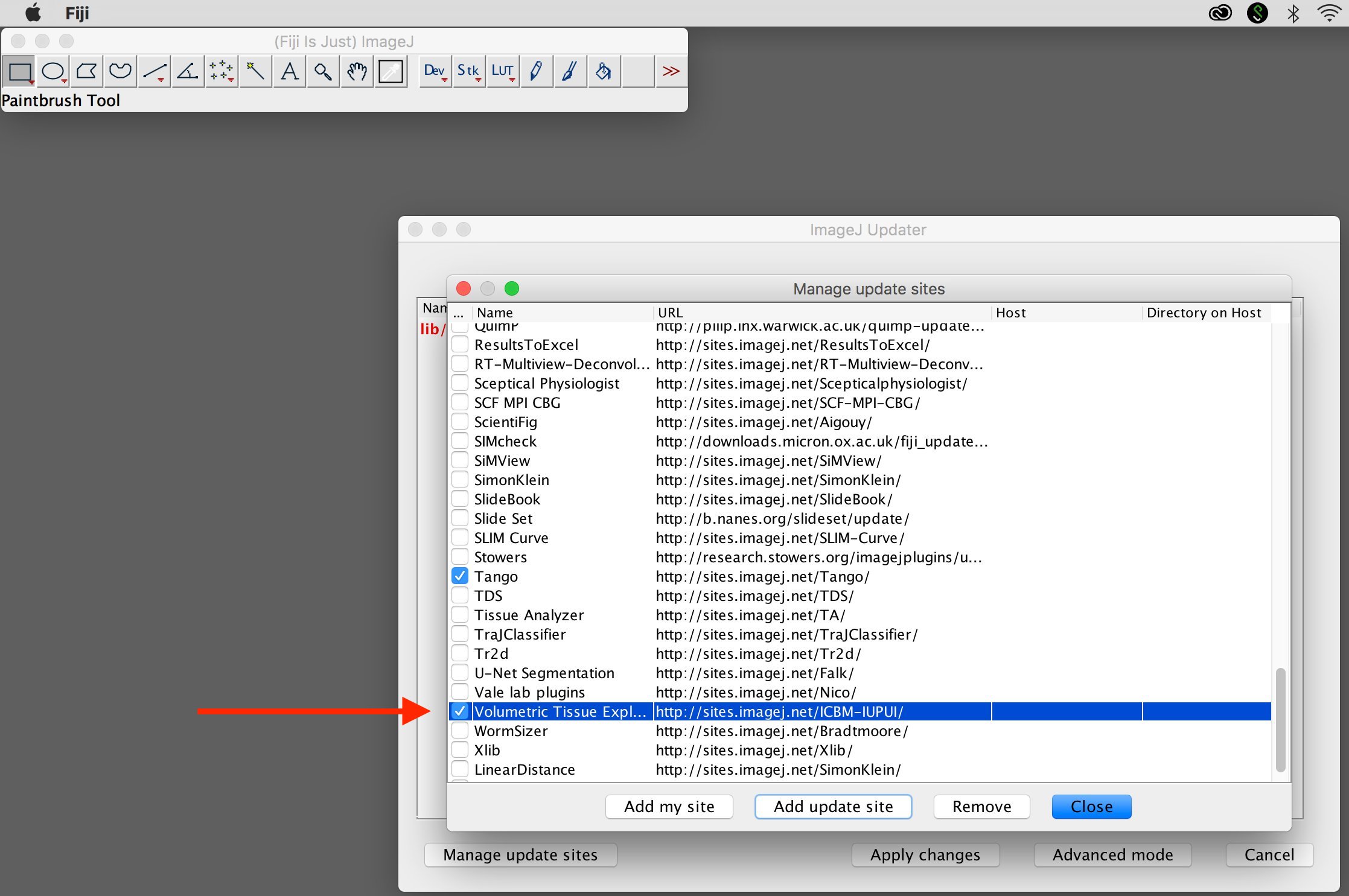
3. Select update site
Check "Volumetric Tissue Expl..." and select "Close"
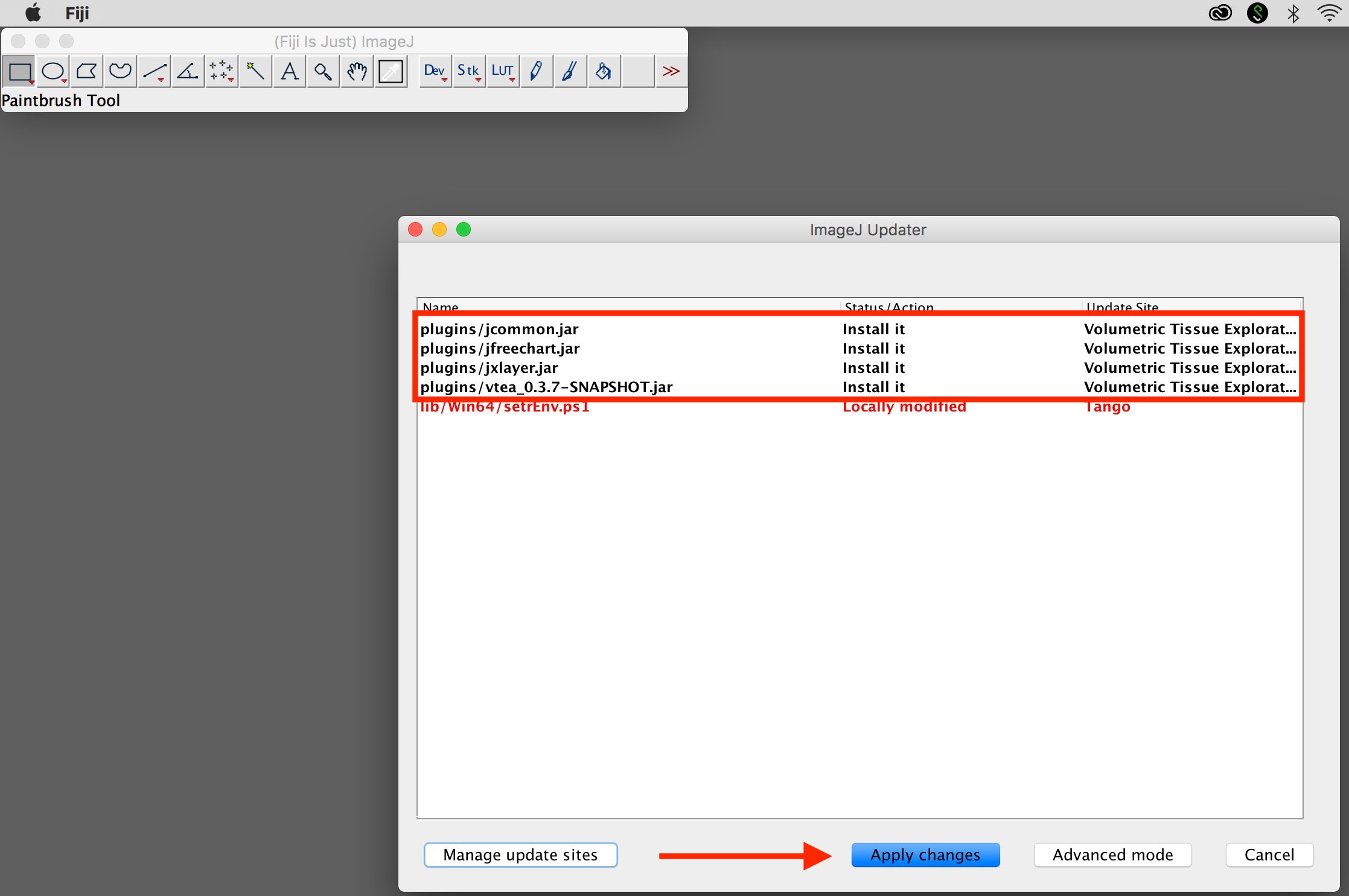
4. Apply changes
Select "Apply changes". Upon restart, the plugin will be present under a new menu "IU_Tools".
Tutorials
Image Processing
Coming soon!
Segmentation
Coming soon!
Analysis
Coming soon!
Visualization
Coming soon!
VTEA reference
The utility of VTEA and 3D tissue cytometry was demonstrated in the following publication:
Quantitative Three-Dimensional Tissue Cytometry to Study Kidney Tissue and Resident Immune Cells Seth Winfree, Shehnaz Khan, Radmila Micanovic, Michael T. Eadon, Katherine J. Kelly, Timothy A. Sutton, Carrie L. Phillips, Kenneth W. Dunn, and Tarek M. El-Achkar JASN ASN.2016091027; published ahead of print February 2, 2017, doi:10.1681/ASN.2016091027