DeformationTracker
| Deformation Tracker | |
|---|---|
| Project | Fiji |
| URL | https://imagej.net/DeformationTracker |
| Source | on GitHub |
| License | GPLv3 |
| Date | Wed Jul 19 12:00:45 CDT 2017 |
| Development status | Active |
| Support status | Active |
| Team | |
| Founders | Jean Leon Maitre, Ozge Ozguc, Varun Kapoor |
| Leads | Varun Kapoor |
| Developers | Varun Kapoor |
| Debuggers | Varun Kapoor, Ozge Ozguc, Andrew Clark |
| Reviewers | Varun Kapoor |
| Support | Varun Kapoor |
| Maintainers | Varun Kapoor |
| Contributors | Varun Kapoor, Ozge Ozguc, Andrew Clark |
ETrack is a tool, which computes the deformation of elliptically shaped deformed cells such as Embryo cells, imaged by either bright-field or fluorescent microscopy.
For using ETrack you have to provide a superpixel segmentation map of your cells in the timelapse image along with the original timelapse images coming out of the microscope.
Contents
Installation
- Click Help ▶ Update....
- Click the Manage update sites button.
- Select the ETrack update site in the list.
- Click Close and then click Apply changes.
- Restart Fiji.
- Launch the plugin with Plugins ▶ ETrack.
Usage
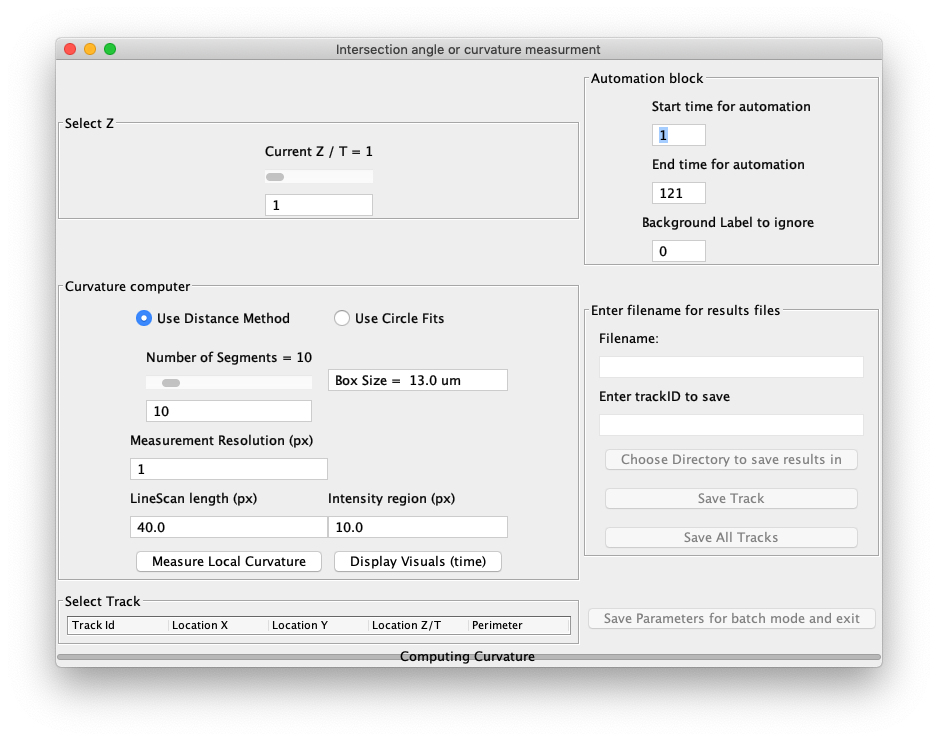
Automated Curvature Measurement
A typical dataset consists of a single two-dimensional (2d) image of the cells whose surface is oscillating over time. The file format can be any format readable by Fiji/Bioformats (.tif, .nd2, … ). To run the tracker select Plugins › ETrack › Automated angle and curvature measurement
The welcome panel will open.
Choose Mode
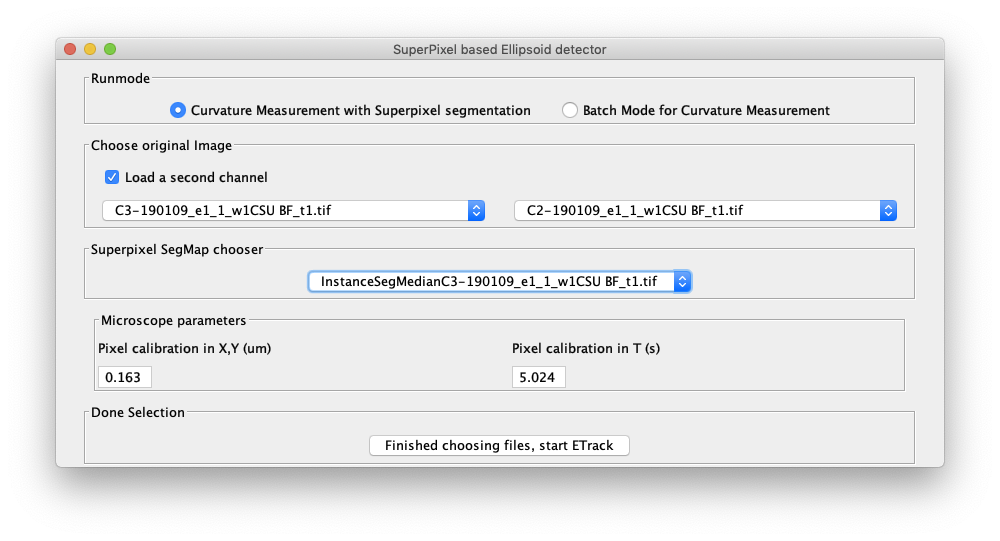
For a first analysis of your data, we suggest using the Curvature Measurement with Superpixel segmentation, in which we have pre-selected a number of parameters. The timelapse images and the segmentation image described below must be opened in Fiji before starting ETrack.
The user has the option of either running the calculation on a single channel or two channels. For a single channel calculation please select an open timelapse image from the dropdown menu. For a double channel calculation select "Load a second channel option" and another drop down menu appears where a second timelapse image can be selected from the open images.
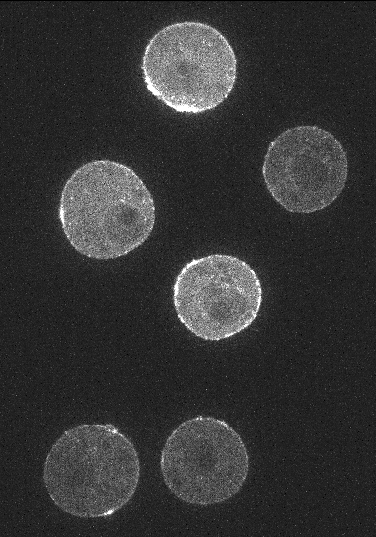
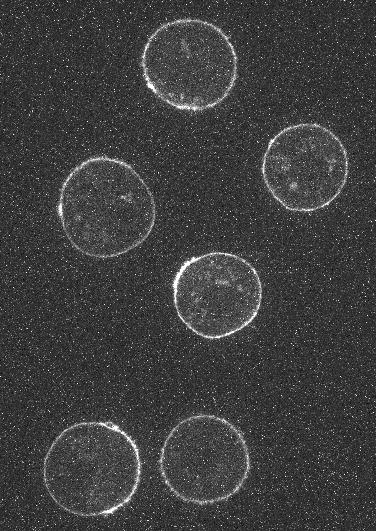
As an example, for Embryo cells the user can select the actin channel image from the left drop down menu and membrane channel image from the right dropdown menu. A segmented image of the cells must also be provided. For a single cell this would be a binary image of the cell and if multiple cells are present in the timelapse image then the segmentation image must be an integer labelled image where each integer label corresponds uniquely to a cell.
Embryo cells with Actin, Membrane channel and segmentation mask image respectively:
Please run the program at least once before running it in batch mode over a directory of such timelapse images.
The microscope parameters are read from the channel 1 image and are displayed in the Pixel calibration boxes in the microscope parameters panel box in the welcome screen (shown above). If the parameters can not be read from the metadata, the user can input the pixel size and timeframe to second conversion.
Press "start ETrack" to proceed. The main ETrack panel will appear and the first time frame of the timelapse image would be displayed. All other open images in Fiji will close. By default, the distance method of deformation measuring tool will run on the first frame of the image.