SPIM Bead Registration
| Selective Plane Illumination Microscopy Bead-based Registration (ImageJ) | |
|---|---|
| Author | Stephan Preibisch, Stephan Saalfeld, Johannes Schindelin, Pavel Tomancak |
| Maintainer | Stephan Preibisch |
| File | SPIM_Registration.jar |
| Initial release | February 2010 |
| Latest version | October 2011 |
| Category | SPIM Registration |
| Website | Stephan Preibisch's homepage |
Contents
Citation
Please note that the SPIM registration plugin available through Fiji, is based on a publication. If you use it successfully for your research please be so kind to cite our work:
- S. Preibisch, S. Saalfeld, J. Schindelin and P. Tomancak (2010) "Software for bead-based registration of selective plane illumination microscopy data", Nature Methods, 7(6):418-419. Webpage PDF Supplement
For technical details about the registration method and SPIM imaging see SPIM Registration Method.
Introduction
Selective Plane Illumination Microscopy (SPIM, Science, 305(5686):1007-9) allows in toto imaging of large specimens by acquiring image stacks from multiple angles. We developed an algorithm for registration of multi-angle SPIM acquisitions using fluorescent beads in rigid mounting medium as fiduciary markers. The beads are matched by invariant, local geometric point descriptors and their displacement is minimized for globally optimal, sample-independent registration. The matching is computed very efficiently using a variation of geometric hashing reducing the matching problem to logarithmic complexity; the computation of the registration for a time point on a single computer takes less time than it takes to acquire the images making it suitable for real-time use. The framework can also be applied to register time-points to each other thus providing optimal alignment of time series.
The developed bead-based registration framework is able to reconstruct long-term, time-lapse, multi-view acquisitions of developing samples with yet unattainable temporal and spatial resolution allowing new insights into the development of biological specimen.
Overview of the SPIM bead-based registration plugin
Please note: the bead-based registration has been rewritten and now replaces the "old" plugin collection (Registration, Advanced Registration, MultiChannel Registration) which has been moved to the deprecated folder. For information regarding these outdated plugins please refer to this page.
The bead-registration plugins offers two registration options:
- Single-channel
- Multi-channel (same beads are visible in different channels)
The typical option is single-channel registration, even if you want to process a multi-channel acquisitions. You can apply the registration of one channel later to other channels in the multi-view fusion plugin. Multi-channel registration is only required if the same beads are visible in several channels.
Both options work in the exactly same way and have very similar options. The additional options of multi-channel registration are explained in the last section.
Important note: the output of the plugin are not images, but only registration matrices that serve as basis for either the multi-view fusion plugin or the multi-view deconvolution plugin. An overview of the complete registration and fusion process can be found here.
How to use the plugin
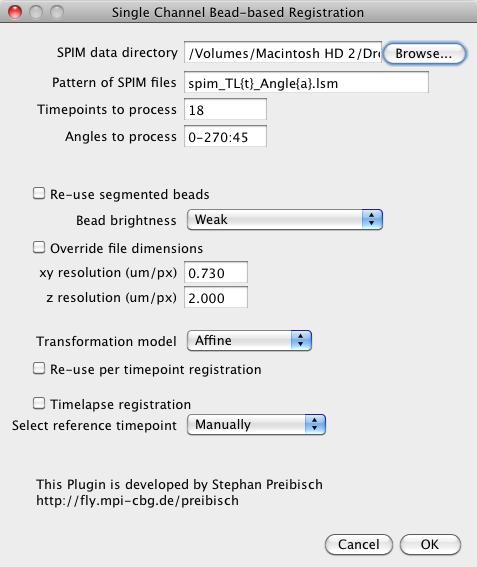
- SPIM data directory: Fill in the directory name that contains all the image files (or directories with equally sized 2d image planes). You can either drag&drop the directory, browse for it or type the name directly.
- Pattern of SPIM files: Define the naming convention of the input files by explaining how the angle and the timepoint is encoded into the filename. For example the file names are named in following way
spim_tl1_angle0.lsm, spim_tl1_angle45.lsm ... spim_tl1_angle270.lsm
spim_tl2_angle0.lsm, ... spim_tl2_angle270.lsm
spim_tl100_angle0.lsm, ... spim_tl100_angle270.lsm.
That means the pattern of the file names corresponds to spim_tl{t}_angle{a}.lsm, where {t} is replaced with the current timepoint and {a} with the current angle. If the numbers contain leading zeros (e.g. 000, 045, 090, 135 instead of 0, 45, 90, 135), this can be encoded by simply adding more letters to the placeholder, in this case {aaa}.
- Timepoints to process: Define the timepoint(s) that should be processed. You can give single numbers (e.g. 18), enumerate (e.g. 1, 18, 19, 100), define ranges (e.g. 1-18) or combine all of them in any combination wanted (e.g. 1,2,10,20-50).
- Angles to process: Define the angles (at least 2) that should be processed (for each timepoint if applicable). You can enumerate angles (e.g. 0, 90, 180, 270), define ranges in steps (e.g. 0-270:45 which means use angle 0 to 270 in steps of 45, i.e. 0, 45, 90, 135, 180, 225, 270) or combine all of them in any combination wanted (e.g. 0, 45, 180-270:45).
- Re-use segmented beads: If beads of all input stacks have been segmented in a previous run, they can be loaded here.
- Bead brightness: If the beads are not loaded, define their approximate brightness, you have the following options:
- Very weak: the beads are hardly or not visible relative to the intensity of the imaged specimen
- Weak: the beads are visible, but not as bright as the sample
- Comparable to sample: the beads are as bright or slightly brighter than the sample
- Strong: the beads are significantly brighter than the sample (you should consider using differently colored beads)
- Advanced...: you know the parameters for the difference-of-gaussian detector, most likely from a previous run of the interactive tool. Note that a new window will pop up to query the parameters
- Interactive...: an interactive tool will allow you to set the approximate parameters for your dataset
- Override file dimensions: If you know the calibration of your dataset you can enter it here and prevent the plugin from opening the file to determine the calibration that is stored (and which might be incorrect)
- Transformation model: Choose the transformation model that will be used to align the datasets to each other, you have three choices:
- Translation: correct for three-dimensional shift
- Rigid: correct for three-dimensional shift and rotation, this option will additionally output an estimate of the rotation axis and rotation angle for each view
- Affine: correct for three-dimensional shift, rotation, shearing and anisotropic scaling, this transformation model best describes the degrees of freedom of SPIM registration
- Re-use per timepoint registration: Loads the registration for each individual timepoint. This is basically only interesting if you perform a timelapse registration (see below).
- Timelapse registration: Checked if a timelapse registration should be performed. In this case the pre-registered individual timepoints are re-registered to one reference timepoint, which results in an aligned time series.
- Select reference timepoint: The reference timepoint can either be selected automatically based on the quality of the individual timepoint registrations or you can choose it yourself (see section timelapse registration).
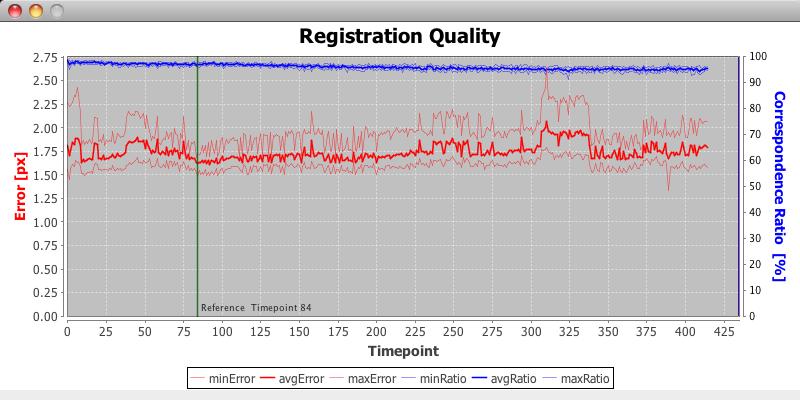
How timelapse registration works
Processing a timelapse acquisition works in two steps. In the first step the registration for each individual timepoint will be computed and automatically stored. Based on these results, a reference timepoint will be selected either manually or automatically (see figure). Note that the panel will be also displayed when choosing automatic reference timepoint selection, but it cannot be changed. Based on the selected timepoint, all other timepoints of the series will be registered relative to it which leads to a properly aligned timeseries.
For fusing the data, the cropping area has to be defined in the reference timepoint. Therefore, the reference timepoint should be fused individually once, and afterwards the whole timeseries can be fused.
Multichannel registration
The multi-channel registration will offer another field in which the channels that contain the same beads can be selected. In the file name pattern channels are coded as {c}, just as timepoints or angles. In the field Channels containing beads you can define the channels to process. You can give single numbers (e.g. 0), enumerate (e.g. 1, 2), define ranges (e.g. 1-3) or combine all of them in any combination wanted (e.g. 1,3-4). The bead brightness for each individual channel will be queried in individual dialogs afterwards.
How do I ... Problems, known issues and solutions
Some tips and tricks in the next few paragraphs require to change a static variable in the source code that changes the behavior of the plugin. This is done using the script editor and works as follows:
- File › New › Script
- Language › Beanshell
- type the command, e.g.
System.gc();
- click Run
This just cleans up the RAM, which might be useful. Note that changes that are made (not in this case though) are only valid for the currently running Fiji instance.
How to set the min and max intensity for each view
The min and max intensity for each view is determined by looking through all the pixels of the stack and finding the lowest and highest intensity. All intensities are then normalized to [0...1] depending on those intensities. If there is a huge difference between different views (maybe because of a piece of dirt only visible in some views), it can be set manually.
mpicbg.spim.registration.ViewDataBeads.minmaxset = new float[]{ 0, 65535 };
This command sets the min and max of all views to 0 and 65535.