Correcting drift in FRAP experiments
This tutorial is brought to you by Joao Firmino, Knust lab, MPI-CBG. It relates how to correct for the drift of your biological samples during long-term timelapse imaging for subsequent analysis using Fiji. Comments on the content of this tutorial are welcome in the disscussion page.
Fluorescence Recovery After Photobleaching in Drosophila embryos
At the confocal
The following tutorial has been optimized to Drosophila embryos using the DuoScan confocal system in the MPI-CBG. Optimization of the bleaching and acquisition parameters while using other model organisms is strongly recommended.
I normally use GFP tagged proteins, so the acquisition laser used is the Argon 488nm and for the bleaching I use the 405nm diode. I set up the Argon laser output power to 45% so that the current through it is around 6A. For the acquisition itself, I set the laser at 10%. In the channel view, I take sections of 2μm (this can be adjusted by the pinhole) and usually slide the gain to around 750. The bleaching conditions are as follow: 488nm laser at 100% and 405nm laser at 100% also. The number of iterations used is 7 - this is probably one of the most important aspects of the experiment: depending on the protein, model organism and size of the region of interest, this number should be increased (this might result in ablation) or decreased (which can cause no bleach at all) according to your needs.
Since you are interested in obtaining a good description of the fluorescence recovery it is important to get the maximum number of time points immediately after the bleach. This is crucial for the aspect of your fitting curve. Therefore you have to get quick time frames with a good enough resolution for your further analysis and quantification. I usually take 512x512 frames with a zoom of 3 and time of acquisition around 1s.
Now you have to establish the duration of the experiment - this depends on the kinetics of recovery of every protein so optimization is advised. In my particular case, every experiment lasts for 21m25s. However there are different acquisition rates during this time. The first 5 frames are taken every 5s without any bleach - this will be your prebleach conditions (25s). After this initial time series, you bleach your sample using the conditions previously described. Immediately after the bleach you take 60 frames every second - this will be your fast acquisition postbleach time series (1m). After this minute of intense imaging you can delay your frame rate to a medium level (one frame every 5s for 10 minutes) - medium acquisition postbleach. And finally you can take frames every 30s for another 10minutes - slow acquisition postbleach. All this can be set up automatically by using the Visual Macro option in the DuoScan confocal system. Once everything is done you should have 4 different time series and 205 frames in total. It is now time to analyze the results...
Analysing the data
Load the 4 different time series in Fiji.
The next step will be to unite the 4 movies in a single one. For this I use the Concatenator plugin for ImageJ which can be found here (https://imagej.net/plugins/concatenator.html). Once you have done this, you should carefully look at your specimen. If it has moved during image acquisition, so will have your bleaching ROI. You can minimize this by using the Linear Stack Alignment with SIFT plugin designed by Stephan Saalfeld which basically compares a frame with its previous one and tries to compensate eventual movement by rigidly moving the whole image so that they better overlap. This way you are not playing around with the fluorescence intensity values and you do not have to manually adjusting your ROI frame by frame. Of course, this plugin does not always work - if your sample moved a lot or if the signal does not allow for efficient comparison, the resulting movie will be pointless.
Once you have an aligned movie you can draw the bleaching ROI and run Plot Z-axis profile (under Images › Stacks). This will not only show you a graph with the mean intensity values over time but will also open a measurement window with raw data. Copy these values to an excel spreadsheet. You still have to normalize these values - the acquisition of images also bleaches somehow the sample and this effect has to be minimized. For that, all you need to do is draw a non-bleached ROI and take out all its values using the above mentioned method.
You can normalize your values by using the following equation:
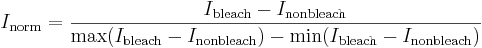
You will get values between 0 and 1 already normalized. All you have to do now is to plot your normalized values against time and you will get a rough curve of recovery. If you want to further analyse this you will have to fit this curve... For this, you can always talk to the Image Processing Facility in house, or if you already have a good knowledge of MATLAB do it yourself...
