Multiple Images (Stacks)
NIH Image has the capability of handling stacks of images, such as those making up
a three dimensional data set.
Boron stack.pics
is an example of a stack of 50
Boron images
(Gillen, 1994)
of grain boundaries in a steel sample. The images
were taken at equal time intervals with an ion microscope, which etches away the surface at
a more or less constant rate. The images thus approximate serial sections of the
steel sample. (The stacks can be saved in two formats - PICS and TIFF. The boron stack requires 618K in PICS format and 4232K in TIFF format. The TIFF format is more widely used, but the PICS format was used
for this example to save space.)
Load the stack using the file - open
command. (This will require more memory than the 4000K recommended for all the preceding
steps. With 4000K of memory assigned to NIH Image, only 13 of the 50 slices will
be loaded. The following steps can be performed on this abbreviated stack, however.)
The images will be displayed one by one as they are loaded, giving a view going
down through the sample. When loaded, the stack is displayed with the Fire-1 color
table, or thermal scale (Fig. 20). Note the "(1/50)" in the title bar indicating that the image shown is the first
of fifty images in the stack. (Note 3. The white diamond in the title bar indicates
that the image is density calibrated, in this case, because "Invert Pixel Values"
was selected.) The > and Stacks - Next Slice
and Stacks - Last Slice
can be used to view other images in the stack. The options - grayscale
command can be used to view the images as they were originally acquired. Under
some conditions, such as after switching back from the finder with the 'Load System
LUT on switching' option set with the options - preferences
command, the system palette, which is useless for this data set, will be in effect. The
gray scale or thermal scale can be restored using the either the options - grayscale
command or the options - color tables - Fire-1
command.
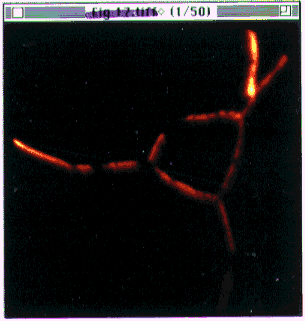
- Figure 20-- The stack of images
(Boron stack.pics)
as it appears after loading.
First image in the stack of 50 is displayed, with a thermal scale. Image dimension
- 100 m.
The stack can be viewed in a variety of ways. The stacks - animate
command cycles through the images, showing them as a movie. The stacks - make montage
command shows images taken in equal intervals throughout the stack as a composite
(Fig. 21).
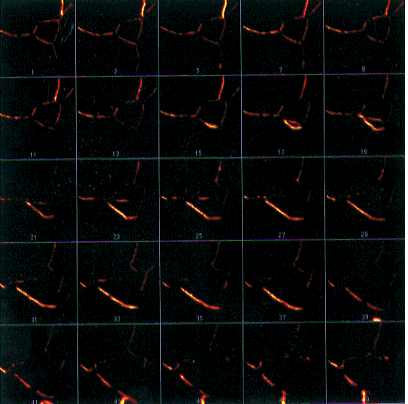
- Figure 21-- Montage of the Boron Stack, with an Increment of 2 in the Stacks -
Make Montage... dialog box.
Stacks can be viewed from other perspectives, treating the layers of the stack as
another spatial dimension. When the number of images in the stack is much less
than the number of pixels along the side of one of the images, the images need to
be replicated to keep the volume as a whole from looking too thin. This can be done by
loading the stacks
macro file (in the Macros folder) with the Special - Load Macros
command. (The macro capability give a powerful way to automate NIH Image. To read
the macro files, open them with NIH Image as a text file (with File - Open)
, or peruse them with another text editor. Programming details can be found in "About
NIH Image" and "Inside Image".) The figures that follow were made with the stack
after using a replication factor of 3 with the newly loaded Special - Replicate Slices
command. This triples the amount of memory required (17 MB assigned in the File - Get Info
dialog box after selecting NIH Image in the finder), but thickens the stack so that
the slice - slice distance approximates the pixel-pixel distance within an image.
The replication of the slices renders the grain structure with the correct scaling
when viewed from the side or from oblique angles. If that amount of memory is not
available, the following steps can still be performed. The aspect ratio of the
flattened images can be corrected using the Edit - Scale and Rotate...
command.
Using the Stacks - Reslice
command, the stack can be viewed in a plane perpendicular to the plane of the images as determined
by the line selection tool (Fig. 22). The resulting image is shown in Fig. 23.
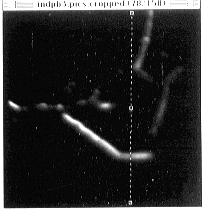
- Figure 22-- Line selection for viewing stack along plane perpendicular to the
images - see next figure.
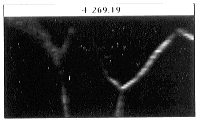
- Figure 23-- Boron stack viewed perpendicular to images in plane intersecting
dotted line, Fig. 22.
The stack can also be rendered as a synthetic volume using the Stacks - Project
command, which shows the images as if they were printed on clear plastic, and stacked
on one another in order to show the three dimensional object as embedded in a plastic
cube. The command generates a new stack of images, each image being a synthesized view of the 'plastic cube' as viewed from a particular angle. The angles
are chosen at equal intervals of rotation about the x, y or z axis so that when
the new Projection stack is animated, the 'plastic cube' appears to rotate, giving
a strong illusion of three-dimensionally. There are a variety of options to this command,
that affect the opacity of the 'plastic' and the structures in the image, all of
which are explained in 'About NIH Image'. Fig. 24 shows the first image, generated
with the default settings, except that 'Brightest point' rather than 'Nearest Point'
was used. Fig. 25 shows a montage of this stack. This operation takes about 2.5
minutes per frame on a Mac IIfx or about 22 seconds per frame on a PowerMac 6100/60. This operation also requires more memory, however there is an automatic option
to store the projected stack on to the disk as separate images when sufficient RAM
is not available. They can then be recombined later into a stack, and animated
to view the grain structure in 'simulated 3-D'.
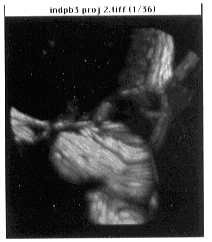
- Figure 24-- Synthetic volume rendering of Boron Stack, done with the Stacks -
Project command (see text). First frame - top view, gray scale.
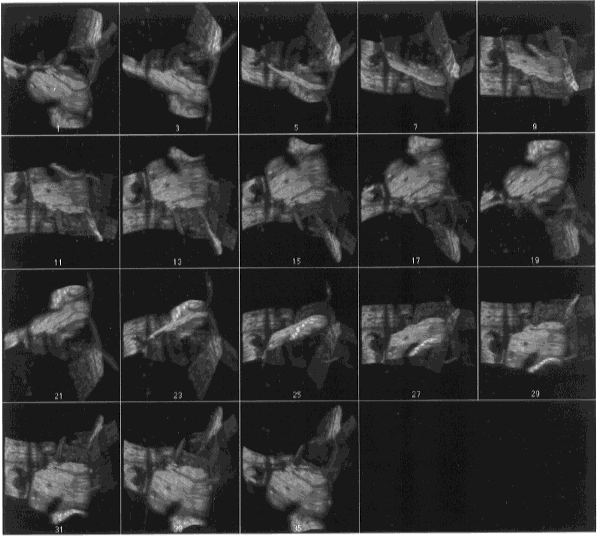
- Figure 25-- Volume rendering of boron stack - montage of every other frame
showing different rotations, gray scale. The three-dimensional structure of these
grain boundaries is more apparent upon viewing the animation of this stack using
the Stacks - Animate command.
[Index]
[Next]






