The content of this page has not been vetted since shifting away from MediaWiki. If you’d like to help, check out the how to help guide!
The ISBI 2012 challenge server has been decommissioned, but you can still download the training and test data!
 The IEEE International Symposium on Biomedical Imaging (ISBI) is the premier forum for the presentation of technological advances in theoretical and applied biomedical imaging and image computing. In the context of the ISBI 2012 conference which was held in Barcelona, Spain, from 2 to 5 May 2012, we organized a challenge workshop on segmentation of neuronal structures in EM stacks.
The IEEE International Symposium on Biomedical Imaging (ISBI) is the premier forum for the presentation of technological advances in theoretical and applied biomedical imaging and image computing. In the context of the ISBI 2012 conference which was held in Barcelona, Spain, from 2 to 5 May 2012, we organized a challenge workshop on segmentation of neuronal structures in EM stacks.
In this challenge, a full stack of EM slices will be used to train machine learning algorithms for the purpose of automatic segmentation of neural structures.
The images are representative of actual images in the real-world, containing some noise and small image alignment errors. None of these problems led to any difficulties in the manual labeling of each element in the image stack by an expert human neuroanatomist. The aim of the challenge is to compare and rank the different competing methods based on their pixel and object classification accuracy.
Relevant dates
- Deadline for submitting results:
February 1stMarch 1st, 2012
- Notification of the evaluation:
February 21stMarch 2nd, 2012
- Deadline for submitting abstracts:
March 1stMarch 9th, 2012
- Notification of acceptance/presentation type:
March 15thMarch 16th, 2012
The workshop competition is done but the challenge remains open for new contributions.
How to participate
Everybody can participate in the challenge. The only requirement consists of filling up the registration form here(offline) to get a user name and password to download the data and upload the results.
This challenge was part of a workshop previous to the IEEE International Symposium on Biomedical Imaging (ISBI) 2012. After the publication of the evaluation ranking, teams were invited to submit an abstract. During the workshop, participants had the opportunity to present their methods and the results were discussed.
Each team received statistics regarding their results. After the workshop, an overview article will be compiled by the organizers of the challenge, with up to three members per participating team as co-authors.
If you have any doubt regarding the challenge, please, do not hesitate to contact the organizers. There is also an open discussion group that you can join here.
Training data
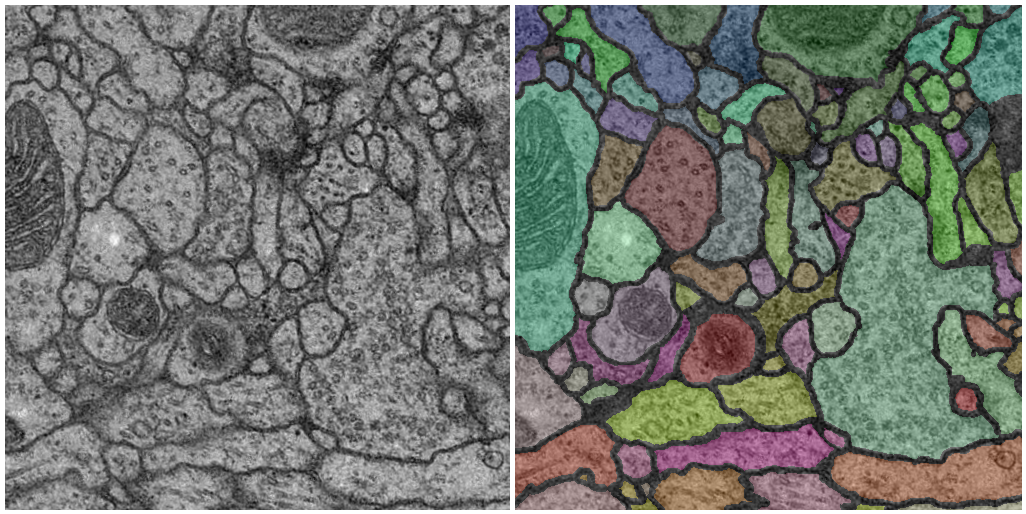
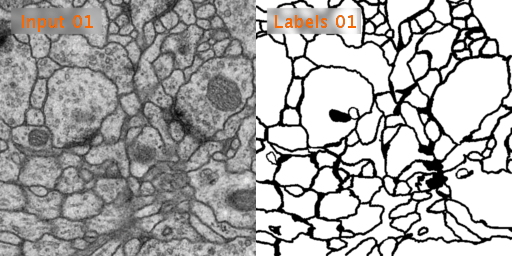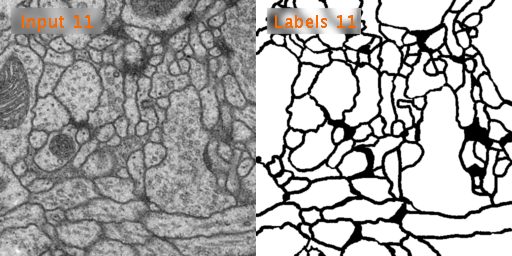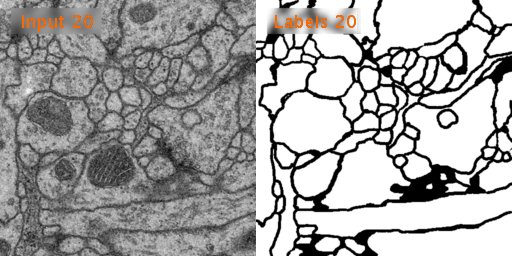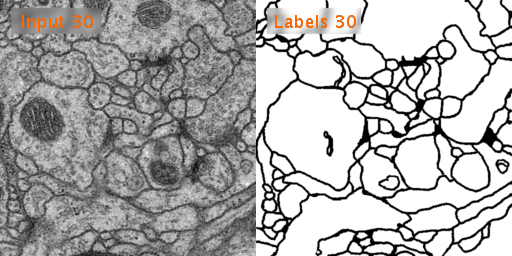
The training data is a set of 30 sections from a serial section Transmission Electron Microscopy (ssTEM) data set of the Drosophila first instar larva ventral nerve cord (VNC). The microcube measures 2 x 2 x 1.5 microns approx., with a resolution of 4x4x50 nm/pixel.
The corresponding binary labels are provided in an in-out fashion, i.e. white for the pixels of segmented objects and black for the rest of pixels (which correspond mostly to membranes).
To get the training data, please, register in the challenge server(offline), log in and go to the “/downloads” section.
This is the only data that participants are allowed to use to train their algorithms.
Test data
The contesting segmentation methods will be ranked by their performance on a test dataset, also available in the challenge server(offline), after registration. This test data is another volume from the same Drosophila first instar larva VNC as the training dataset.
Results format
The results are expected to be uploaded in the challenge server(offline) as a 32-bit TIFF 3D image, which values between 0 (100% membrane certainty) and 1 (100% non-membrane certainty).
Evaluation metrics
In order to evaluate and rank the performances of the participant methods, we will use 2D topology-based segmentation metrics, together with the pixel error (for the sake of metric comparison). Each metric will have an updated leader-board.
The metrics are:
- Minimum Splits and Mergers Warping error, a segmentation metric that penalizes topological disagreements, in this case, the object splits and mergers.
- Foreground-restricted Rand error: defined as 1 - the maximal F-score of the foreground-restricted Rand index, a measure of similarity between two clusters or segmentations. On this version of the Rand index we exclude the zero component of the original labels (background pixels of the ground truth).
- Pixel error: defined as 1 - the maximal F-score of pixel similarity, or squared Euclidean distance between the original and the result labels.
If you want to apply these metrics yourself to your own results, you can do it within Fiji using this script.
We understand that segmentation evaluation is an ongoing and sensitive research topic, therefore we open the metrics to discussion. Please, do not hesitate to contact the organizers to discuss about the metric selection.
Organizers
- Ignacio Arganda-Carreras (Howard Hughes Medical Institute, Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge, MA, USA)
- Sebastian Seung (Howard Hughes Medical Institute, Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge, MA, USA)
- Albert Cardona (Institute of Neuroinformatics, Uni/ETH Zurich, Switzerland)
- Johannes Schindelin (University of Wisconsin-Madison, WI, USA)
References
Publications about the data:
doi:10.1371/journal.pbio.1000502
Publications about the metrics: